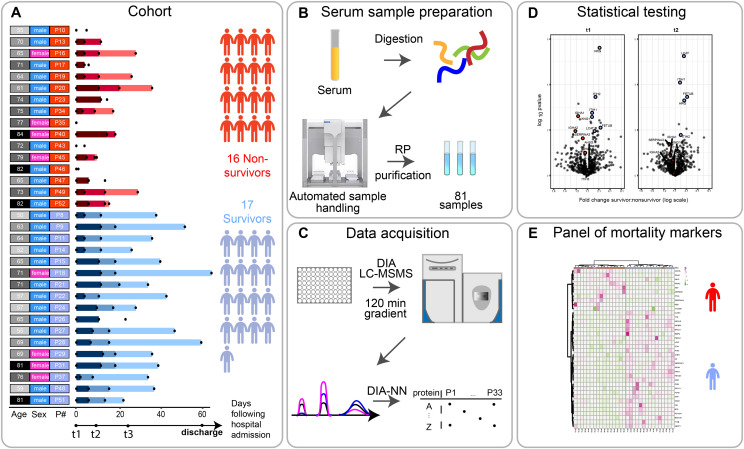
Figure 1. Scheme of the cohort and timing of the blood sample collection, based on each patient’s admission to the hospitalization.
(A) Serum samples were collected from 33 individuals (17 survivors, 16 deceased) diagnosed with SARS-CoV-2 infection, at one, two, or three time points after their admission to the clinic (t0). The time points t1, t2, and t3 represent blood collections at 96 h, 14 d, and later than 14 d after they arrived at the ICU of the hospital. The date of discharge from the hospital is recorded here, although no blood was collected at that moment. The numbers represent the elapsed number of days starting from t1 for each patient. These are represented by color gradients ranging from dark to light the longer the duration of the stay in the clinic. Patients where no color timeline is represented, indicate cases for which no consecutive temporal collection points were available. Patient descriptors including age and gender as well as indexes used throughout this report are provided to the right of the timelines and color coded. Patient’s age is binned (10 yr/bin) and the darker the greyscale in column 1 the older the patient. The gender of each patient is marked in column 2 as blue and pink. Patient indexes are color-coded for patient outcomes, with survivors in blue and deceased SARS-CoV-2 patients in red in column 3. (B) Serum samples were proteolytically digested and the resulting tryptic peptides were purified using reverse-phase cartridges on an autosampler robot. (C) The samples were analyzed by liquid chromatography tandem mass spectrometry (LC-MS/MS) applying a data-independent acquisition strategy. Spectra were extracted using DIA-NN yielding as a measure abundance for each protein. (D) Two-sided t tests were performed to determine significantly regulated proteins comparing survivors and non-survivors. (E) These differentially regulated proteins were found to be largely functionally related, and define a potential panel of mortality markers, by which we can stratify patients that might ultimately overcome or succumb from the infection, which can be diagnosed already at an early time point in the clinic.