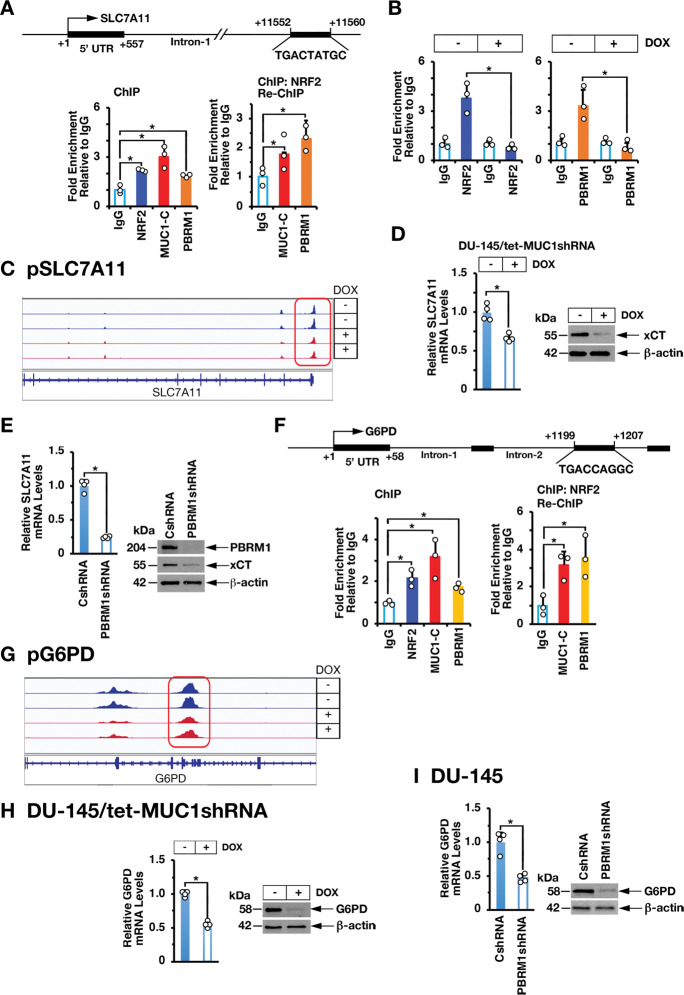
Fig. 5. MUC1-C and PBRM1 interact with NRF2 to activate SLC7A11 and G6PD expression.
A Schema of the SLC7A11 promoter region with highlighting of the NRF2 binding site in intron-1. Soluble chromatin from DU-145 cells was precipitated with anti-NRF2, anti-MUC1-C, anti-PBRM1 or a control IgG (left). Soluble chromatin from DU-145 cells was precipitated with anti-NRF2 (ChIP) and then reprecipitated with anti-MUC1-C, anti-PBRM1 or a control IgG (re-ChIP). B Soluble chromatin from DU-145/tet-MUC1shRNA cells treated with vehicle or DOX for 7 days was precipitated with anti-NRF2 (left), anti-PBRM1 (right) or a control IgG. The DNA samples were amplified by qPCR with primers for the SLC7A11 promoter region. The results (mean ± SD of 3 determinations) are expressed as fold enrichment relative to that obtained with the IgG control (assigned a value of 1). C Chromatin from DU-145/tet-MUC1shRNA cells treated with vehicle or DOX for 7 d was analyzed for ATAC-seq. UCSC genome browser snapshot of ATAC-seq data from the SLC7A11 gene showing loss of peaks and decrease in chromatin accessibility as a function of MUC1-C silencing. D and E. DU-145/tet-MUC1shRNA cells treated with vehicle or DOX for 7 days (D) and DU-145/CshRNA and DU-145/PBRM1shRNA cells (E) were analyzed for SLC7A11 mRNA levels by qRT-PCR. The results (mean ± SD of 4 determinations) are expressed as relative mRNA levels compared to that obtained for CshRNA cells (assigned a value of 1)(left). Lysates were immunoblotted with antibodies against the indicated proteins (right). F Schema of the G6PD promoter region with highlighting of the NRF2 binding site in intron-2. Soluble chromatin from DU-145 cells was precipitated with anti-NRF2, anti-MUC1-C, anti-PBRM1 or a control IgG (left). Soluble chromatin from DU-145 cells was precipitated with anti-NRF2 (ChIP) and then reprecipitated with anti-MUC1-C, anti-PBRM1 or a control IgG (re-ChIP)(right). The DNA samples were amplified by qPCR with primers for the G6PD promoter region. The results (mean ± SD of 3 determinations) are expressed as fold enrichment relative to that obtained with the IgG control (assigned a value of 1). G Chromatin from DU-145/tet-MUC1shRNA cells treated with vehicle or DOX for 7 d was analyzed for ATAC-seq. UCSC genome browser snapshot of ATAC-seq data from the G6PD gene showing loss of peaks and decrease in chromatin accessibility as a function of MUC1-C silencing. H and I. DU-145/tet-MUC1shRNA cells treated with vehicle or DOX for 7 days (H) and DU-145/CshRNA and DU-145/PBRM1shRNA cells (I) were analyzed for G6PD mRNA levels by qRT-PCR. The results (mean ± SD of 4 determinations) are expressed as relative mRNA levels compared to that obtained for CshRNA cells (assigned a value of 1) (left). Lysates were immunoblotted with antibodies against the indicated proteins (right).