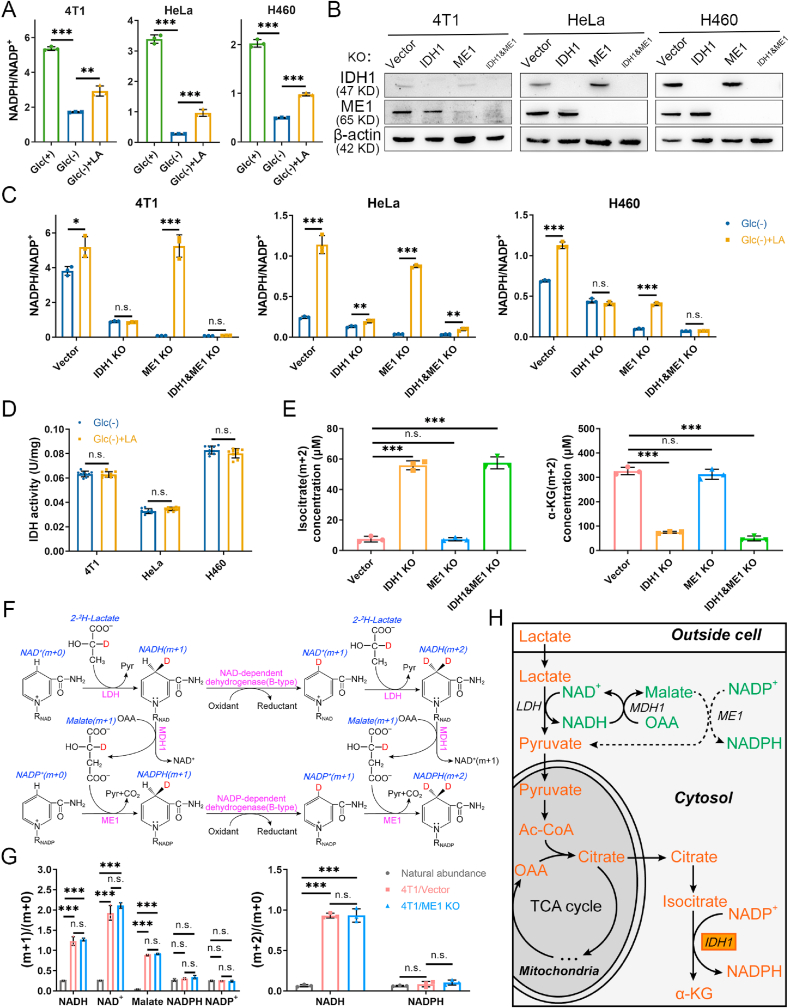
Fig. 1.
Lactate supports NADPH generation by IDH1 rather than ME1 under glucose-deprived condition with lactic acidosis. (A) Cellular NADPH/NADP+ ratios of three cancer cell lines under three different conditions. Incubation times were 9 h (4T1), 3 h (HeLa) and 6 h (H460), respectively. (B) Confirmation of IDH1 knockout, ME1 knockout, and double gene knockout in three cancer cell lines by immunoblot. The cells transfected with empty px459 vector were used as knockout controls. (C) The effect of lactic acidosis on NADPH/NADP+ ratios of IDH1 or/and ME1 knockout cancer cells under glucose-deprived condition. Incubation times were 6 h (4T1), 3 h (HeLa) and 4 h (H460), respectively. (D) The effect of lactic acidosis on total IDH activities of the three cancer cells. (E) Concentrations of isocitrate (m+2) and α-KG (m+2) after labeling with 20 mM [U–13C3]-lactate for 6 h in 4T1 knockout cells. (F) Theoretical pathways to transfer electron pairs from C-2 of lactate to the nicotinamide ring of NADPH. Except the nicotinamide ring, the remaining moiety of NAD(P) is denoted by RNAD(P). (G) The m+1 (2)/m+0 ratios of the intermediates in the electron transfer pathways after labeling with 20 mM [2-2H]-lactate for 6 h in 4T1 cells. The labeling data were not corrected for natural isotopic abundance. (H) Schematic diagram to illustrate NADPH maintenance by lactate under glucose-deprived conditions. The fluxes indicated by dash lines are marginal. Glc (+): Complete RPMI-1640 medium; Glc (−): Glucose-free RPMI-1640 medium; Glc (+) + LA: Glucose-free RPMI-1640 medium supplemented with 20 mM lactic acid (4T1 and HeLa, pH 6.7) or with 20 mM lactic acid and 10 mM NaOH (H460, pH 7.1). Data are shown as means ± SD, with n = 3 biological replicates in (A), (C), (E) and (G), and the results were all confirmed by three independent experiments; Data are means ± SD, from three independent experiments performed in triplicates in (D). n. s. not significant, *p < 0.05, **p < 0.01, ***p < 0.001 (Student's test). See also Supplementary Figs. 1–3.