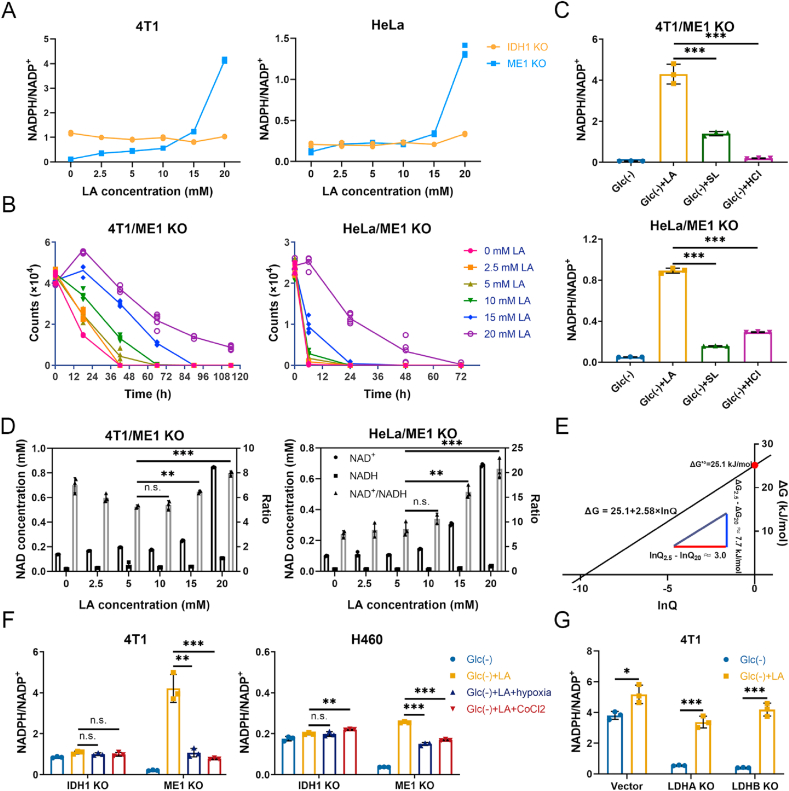
Fig. 4.
Lactate-involved NADPH production is constrained by the thermodynamic barrier of LDH-catalyzed reaction. (A) The effect of lactate concentrations on cellular NADPH/NADP+ ratios of IDH1 or ME1 knockout cells under glucose-deprived condition. (B) The effect of lactate concentrations on the survival of ME1 knockout cells under glucose-deprived condition. (C) Cellular NADPH/NADP+ ratios of ME1 knockout cells under four different conditions: glucose-free, glucose free with lactic acidosis, glucose free with lactosis, and glucose free with acidosis. (D) The effect of lactate concentrations on cellular NAD+, NADH concentrations and NAD+/NADH ratios of ME1 knockout cells under glucose-deprived condition. (E) The linear relationship between ΔG and ln Q of the LDH-catalyzed reaction. The red line means the difference of ln Q between 2.5 mM and 20 mM lactate, the blue line means the corresponding difference of ΔG. (F) Effect of hypoxia (1% O2) or CoCl2 (200 μM) on cellular NADPH/NADP+ ratios of IDH1 or ME1 knockout cells under glucose deprivation with lactic acidosis. (G) The effect of lactic acidosis on cellular NADPH/NADP+ ratios of LDHA or LDHB knockout cancer cells under glucose-deprived condition. The ratios of 4T1 control here are the same with that of Fig. 1C, because the two experiments were in fact performed simultaneously. Glc (−): glucose-free RPMI-1640 medium; Glc (−) + LA: glucose-free RPMI-1640 medium supplemented with 20 mM lactic acid, pH 6.7. Glc (−) + SL: glucose-free RPMI-1640 medium supplemented with 20 mM sodium lactate, pH 7.4. Glc (−) + HCl: glucose-free RPMI-1640 medium supplemented with 20 mM hydrochloric acid, pH 6.7. Glc (−) + LA + hypoxia: glucose-free RPMI-1640 medium supplemented with 20 mM lactic acid under hypoxia (1% O2) condition. Glc (−) + LA + CoCl2: glucose-free RPMI-1640 medium supplemented with 20 mM lactic acid and 200 μM CoCl2. Incubation times were 6 h (4T1), 3 h (HeLa) and 4 h (H460), respectively. Data are shown as means ± SD, with n = 3 biological replicates in (A), (C), (D), (F) and (G), n = 5 biological replicates in (B), and the results were all confirmed by three independent experiments. n. s. not significant, *p < 0.05, **p < 0.01, ***p < 0.001 (Student's test). See also Supplementary Fig. 6. . (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)