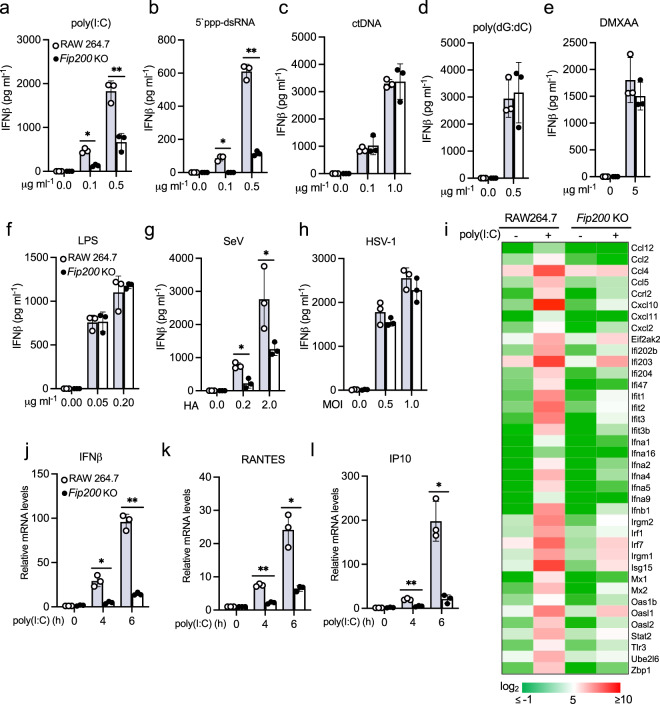
Fig. 4. FIP200 deficiency impairs RIG-I activation in macrophages.
a–f Wild type and FIP200 knockout RAW264.7 macrophages were stimulated with the indicated amount of poly(I:C) (a), 5′ ppp-dsRNA (b), calf thymus DNA (ctDNA) (c), poly(dG:dC) (d), DMXAA (e), or LPS (f). After 16 h, the supernatants were collected for IFNβ ELISA assays. All experiments were biologically repeated three times. Data represent means ± s.d. of three independent experiments. The P-value was calculated (two-tailed Student’s t test) by comparison with wild-type cells. *P < 0.05, **P < 0.01. g, h Wild type and the FIP200 knockout RAW264.7 macrophages were infected with SeV (g) or HSV-1 d109 mutant virus (h). After 16 h, the supernatants were collected for IFNβ ELISA assays. All experiments were biologically repeated three times. Data represent means ± s.d. of three independent experiments. The P-value was calculated (two-tailed Student’s t test) by comparison with wild-type cells. *P < 0.05. i Heatmap of the RNA sequencing results of wild type and FIP200 knockout RAW264.7 macrophages stimulated with 1 μg ml−1 poly(I:C) for 4 h. j–l Wild type and FIP200 knockout RAW264.7 macrophages were stimulated with 1 μg ml−1 poly(I:C) for indicated times. Real-time PCR was performed to determine the relative mRNA levels of IFNβ (j), RANTES (k), and IP10 (l). All experiments were biologically repeated three times. Data represent means ± s.d. of three independent experiments. The P-value was calculated (two-tailed Student’s t test) by comparison with wild-type cells. *P < 0.05, **P < 0.01.