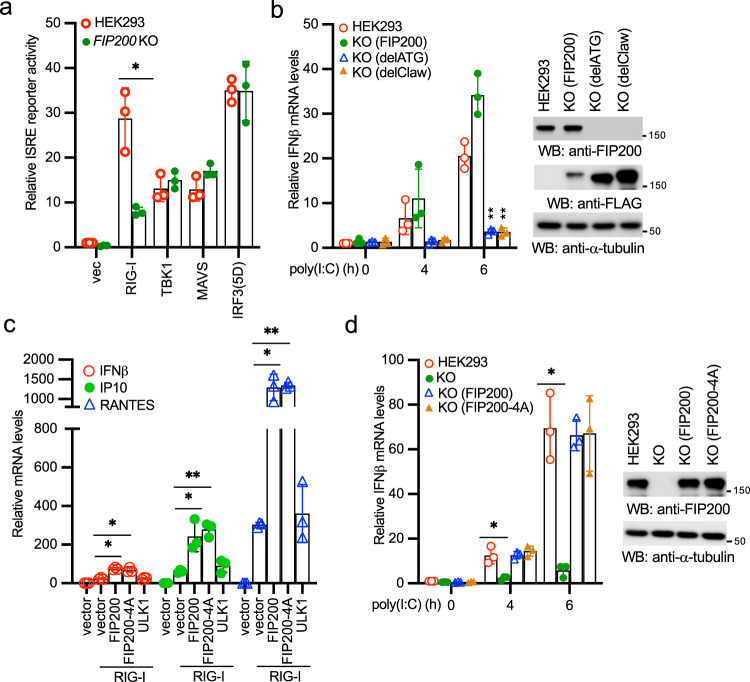
Fig. 5. FIP200 activates innate immunity via RIG-I but independently of its autophagy function.
a FIP200 wild type and knockout HEK293 cells were transfected with vector, RIG-I, TBK1, MAVS, or IRF3(5D) together with pRL-SV40 and pISRE-Luc. After 48 h, cells were collected and the ratio of firefly luciferase to Renilla luciferase was calculated to determine the relative activity of IFN reporter. All experiments were biologically repeated three times. Data represent means ± s.d. of three independent experiments. The P-value was calculated (two-tailed Student’s t test) by comparison with HEK293 cells. *P < 0.05. b Wild-type HEK293 cells, FIP200 knockout cells reconstituted with full-length FIP200, delATG, or delClaw were stimulated with 1 μg ml−1 poly(I:C) for designated times. Then, RNA was extracted, and real-time PCR for IFNβ was performed. All experiments were biologically repeated three times. Data represent means ± s.d. of three independent experiments. (**P < 0.01 vs. the wild-type cells by two-tailed Student’s t test). Right panel shows the expression of FIP200 and the mutants in the reconstituted cells. The anti-FIP200 antibody detects the C-terminal end of FIP200 and cannot recognize delATG and delClaw. c RIG-I was transfected with vector, FIP200, FIP200-4A, or ULK1 into HEK293 cells. After 48 h, RNA was extracted, and real-time PCR assays for IFNβ, IP10, and RANTES were performed. All experiments were biologically repeated three times. Data represent means ± s.d. of three independent experiments. (*P < 0.05, **P < 0.01, by two-tailed Student’s t test). d Wild-type HEK293 cells, FIP200 knockout cells, and FIP200 knockout cells reconstituted with full-length FIP200 or the 4A mutant were stimulated with 1 μg ml−1 poly(I:C) for designated times. Then, RNA was extracted, and real-time PCR for IFNβ was performed. All experiments were biologically repeated three times. Data represent means ± s.d. of three independent experiments. (*P < 0.05 vs. the wild-type cells by two-tailed Student’s t test). Right panel shows the expression of FIP200 and the mutant in the reconstituted cells.