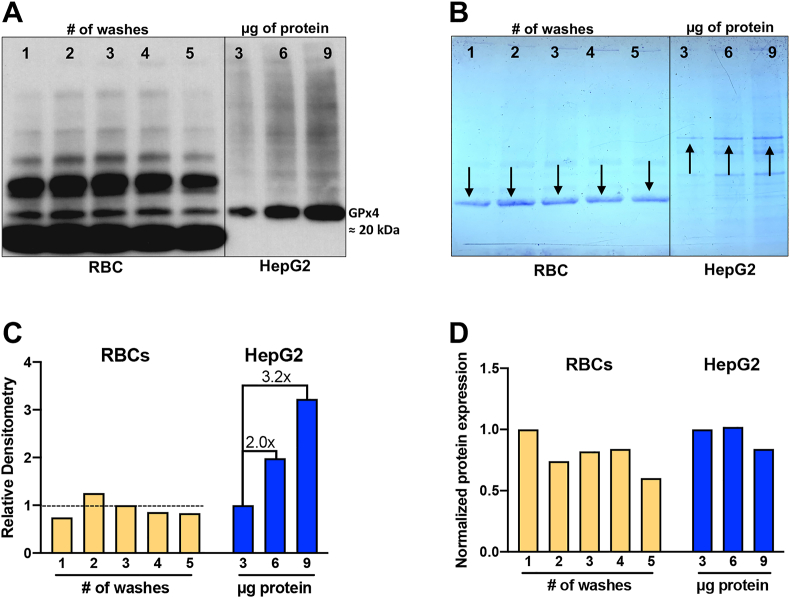
Fig. 4.
Validation of GPx4 antibody and loading control for Western blot analysis of RBCs.
(A) Western blot of samples from RBCs and HepG2 cells. Approximately 4 × 108 RBCs from one subject were washed 1–5 times and 10 μg of protein from each wash was loaded onto the gel. After 3 washes there may be a small loss of GPx4 protein. This is to be expected as GPx4 is non-covalently bound to membranes [33]. HepG2 lysate, as a positive control, shows an increase in GPx4 as more cellular protein is loaded.
(B) The Coomassie-stained gel shows that the intensity of bands corresponds to the amount of protein loaded. Appropriate bands were selected (marked by arrows) for the RBCs and HepG2 cells to perform densitometry measurements, panel C·
(C) Quantification of the Coomassie bands for RBCs shows that the amount of protein loaded in each of the 5 lanes is approximately the same. The Coomassie stain for the HepG2 samples shows the expected increase in band density with increasing amounts of protein loaded (i.e., 3, 6, and 9 μg of protein loaded would be expected to have relative intensities of 1x, 2x, and 3x, respectively). The observed intensities are as anticipated.
(D) The intensity of GPx4 bands (panel A) were normalized to the intensities of the Coomassie-stained bands. An apparent decrease of RBC GPx4 was observed with increasing numbers of washes. Because increasing amounts of protein were loaded for the HepG2 positive control samples, the relative GPx4 expression is similar for all three samples, as expected.