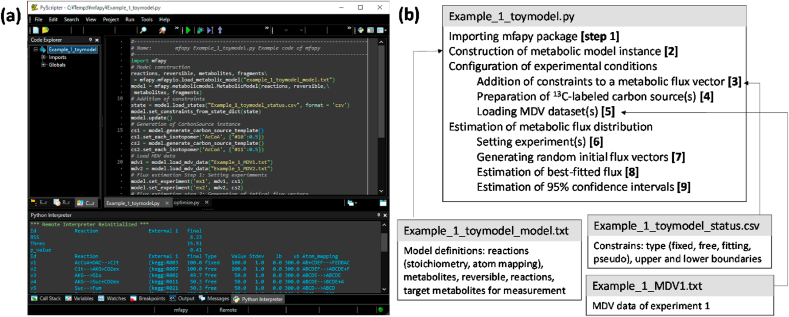
Fig. 1.
Procedure for 13C-MFA by mfapy. (a) Screenshot of a python environment (PyScripter) running mfapy. (b) A typical scheme for estimating a best fitted metabolic flux distribution using the model definition, status, and MDV data files. Python codes for the parallel labeling experiment and the metabolic network of the toy model are shown in Supplementary Figs. 1 and 2.