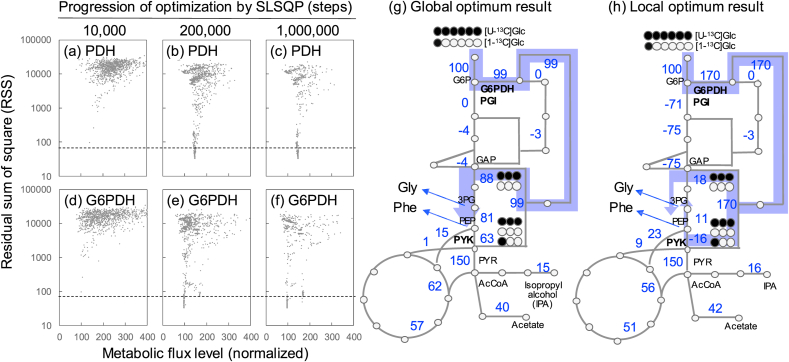
Fig. 3.
Investigation of global and local optima in 13C-MFA of metabolically engineered E. coli. Metabolic models and measurement data were obtained from a previous study (Okahashi et al., 2017). Model fitting was performed by the gradient-based local optimization (SLSQP). A total of 1000 optimization trials were executed in parallel. (a–f) Progression of optimization of 1000 trials. Metabolic flux levels of PDH (a–c) and G6PDH (d–f) reactions at the 10,000th (a and d), 200,000th (b and e), and 1,000,000th (c and f) steps are shown in figure. (g and h) Metabolic flux distribution of global (g) and local (h) optimum results. Blue numbers represent metabolic flux levels. Blue lines indicate significant carbon flow toward glycine and phenylalanine. White and black circles indicate 13C-labeling patterns of carbon source (glucose, Glc) and GAP and PYR produced by the ED pathway. All metabolic flux levels are normalized to that of the glucose uptake rate. (For interpretation of the references to colour in this figure legend, the reader is referred to the Web version of this article.)