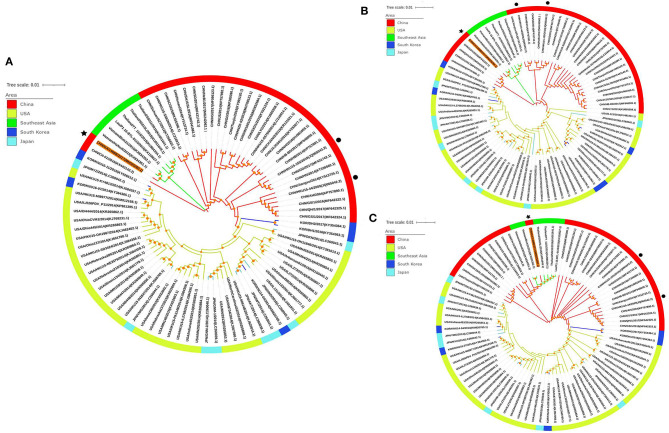
Figure 2.
Phylogenetic trees of PDCoV strains based on the sequences of the complete genome (A) and the S protein (B) and ORF1a/1b proteins (C). Phylogenetic trees were constructed using the neighbor-joining method in MEGA6 software and a bootstrap analysis was performed with 1,000 replicates. The trees were visualized using the online software, iTOL (https://itol.embl.de/), and bootstrap values >50% are shown at the branch points. Each PDCoV strain is indicated in the following format: country origin (CHN, China; JPN, Japan; KOR, South Korea; USA, the United States; Lao; Thailand and Vietnam)/strain name/year of collection (GenBank accession number). The CHN/GX/1468B/2017 strain obtained in this study is indicated with a black asterisk, the CHN/HKU15-44/2009 and CHN/HKU15-155/2010 strains originally identified in Hong Kong are indicated with a black dot. The branches in red, green, yellow, light blue, and dark blue represent the clades from China, Southeast Asia, USA, Japan, and South Korea, respectively.