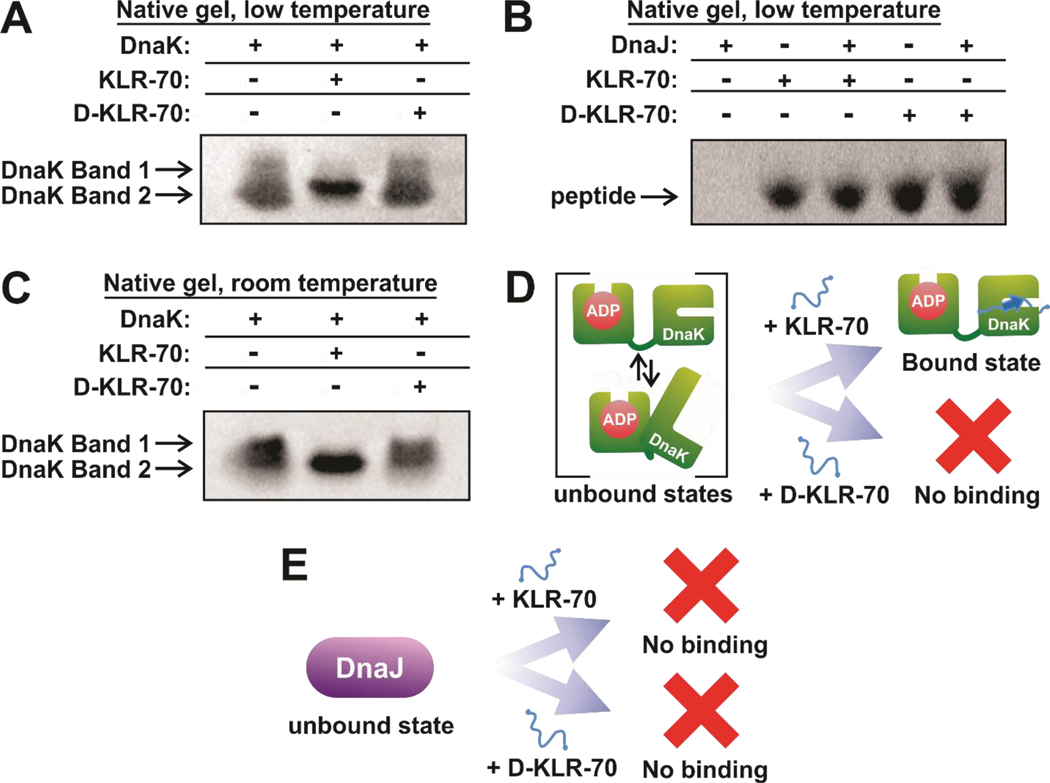
Figure 4.
KLR-70 enantioselectively binds DnaK and does not bind DnaJ. (A) Representative native gel data, run under conventional polarity and on ice (15.0 – 24.5 ± 0.5 °C), for the binding of DnaK (5 μM) to either KLR-70 or D-KLR-70 (50 μM each). (B) Representative native gel data, run under reverse polarity and on ice (15.0 – 24.5 ± 0.5 °C), for the binding of DnaJ (200 μM) to either KLR-70 or D-KLR-70 (35 μM each). A one-tailed Student T-test assuming unequal variances shows that the gel band intensities for the KLR-70 (p = 0.32) and D-KLR-70 (p = 0.39) peptides in the presence of DnaJ are not statistically different. A 95 % confidence interval cutoff (p = 0.05) was used. Native PAGE analysis of these samples at pH 6.8, to allow DnaJ entering the gel, confirm the lack of interaction with DnaJ (Figure S1). (C) Representative native-gel data, run under conventional polarity at room temperature (21.5 ± 0.5 °C), for the binding of DnaK (5 μM) to either KLR-70 or D-KLR-70 (50 μM each). (D) Schematic cartoon representation of the DnaK-peptide binding data in panels A and C. Unbound DnaK is conformationally heterogeneous and it adopts a single apparent conformation upon interaction with KLR-70. In contrast, DnaK does not interact with D-KLR-70. (E) Schematic cartoon representation of the DnaJ-peptide binding data in panel B. Neither KLR-70 nor D-KLR-70 binds DnaJ. All native gel data are representative of n = 3 independent experiments. Native gel electrophoresis was carried out at pH 7.5 (see Material and Methods for additional information).