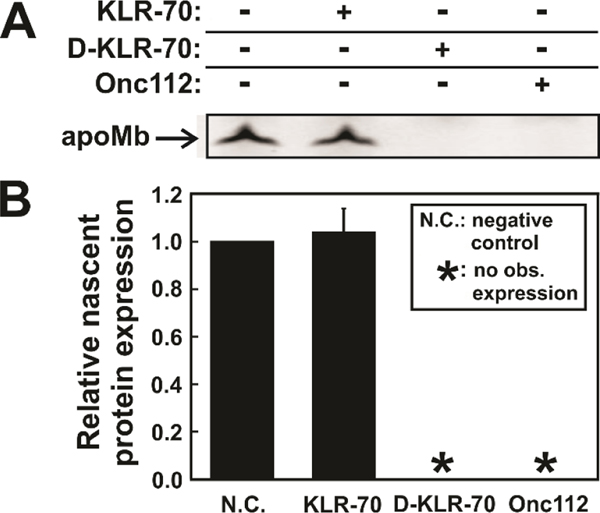
Figure 8.
KLR-70 and D-KLR-70 have a dramatically different effect on cell-free protein expression. (A) Representative low pH SDS-PAGE gel illustrating the cell-free expression of fluorophore-labeled apoMb in the absence and presence of the KLR-70, D-KLR-70 and Onc112 peptides. (B) Quantitative analysis of nascent-protein expression data of panel A, in the absence (N.C.; negative control) and presence of DnaK-binding peptides. Nascent protein expression is quantified relative to expression level of the N.C. sample. Data are shown as avg ± std. err. for n = 3 independent experiments carried out with an E. coli cell-free system lacking the gene for the trigger factor chaperone (Δtig strain). The total concentration of the K/J/E chaperones in the Δtig S30 cell extract were 0.7 μM DnaK, 0.03 μM DnaJ and 0.07 μM GrpE. Asterisks (*) denote samples where no observable (obs.) nascent-protein expression could be detected.