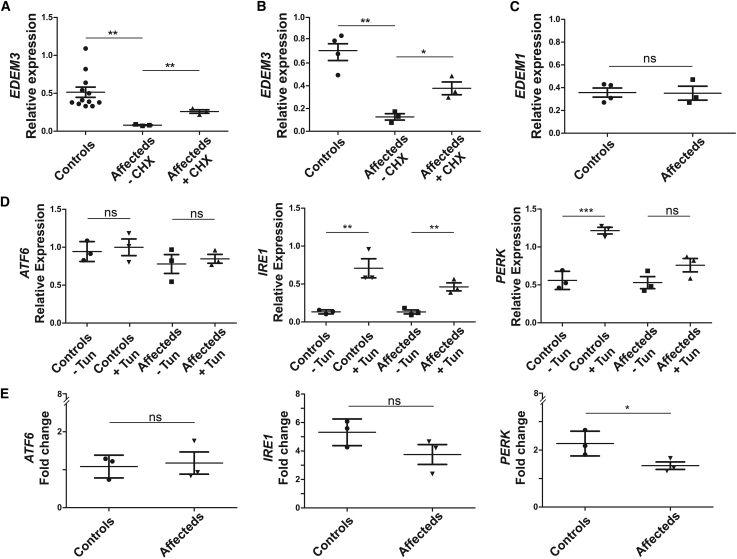
Figure 2.
Relative expression of EDEM3 mRNA and expression analysis of UPR markers
(A) A significant decrease of mRNA levels is seen between relative expressions of EDEM3 in EBV-LCL from three affected individuals of family 1 with the bi-allelic c.1859del frameshift variant (n = 3) compared to healthy controls (n = 12; p = 0.0078) and samples treated with cycloheximide (CHX; n =3; p = 0.0016), normalized according to three housekeeping genes.
(B) EDEM3 expression in fibroblast cell lines from family 1 (individual IV-4) and family 3 (individuals II-1 and II-2) confirming these results (p = 0.0015 and p = 0.0161 for CHX). Four controls were used.
(C) EDEM1 expression in fibroblast cell lines from family 1 (individual IV-4) and family 3 (individuals II-1 and II-2) showing normal expression levels (p = 0.9373). Four controls were used.
(D) Analysis of total mRNA from EBV-LCL cell lines from controls revealed increased mRNA expression after treatment with tunicamycin (Tun) of IRE1 and EIF2AK3 (PERK) but not of ATF6.
(E) Affected individuals’ mRNA from family 1 showed significantly decreased induction of PERK expression when compared to controls (p = 0.020), whereas ATF6 and IRE1 did not change significantly (p = 0.423 and p = 0.091, respectively). Experiments in (D) and (E) were performed with three control cell lines and three cell lines from affected individuals. Bars indicate mean values. Error bars represent standard deviation. ns, not significant; ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001.