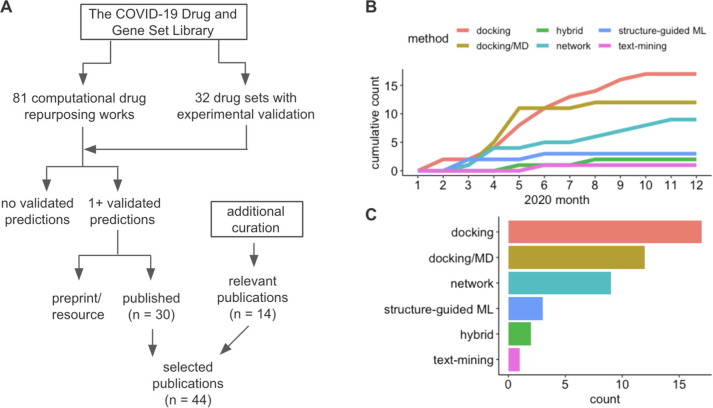
Figure 1.
Selection and classification of works on computational drug repurposing for COVID-19. A. To identify the most effective computational drug repurposing works, we used the COVID-19 Drug and Gene Set Library,35 complemented by manual curation of the literature. The selection criteria aimed to identify methods that resulted in at least one predicted compound that was validated with further experiments. B. We classified the selected works into six groups: docking, docking/MD (molecular dynamics), network, structure-guided machine-learning (ML), hybrid and text mining. The publication trends for these groups in 2020 are shown. C. The final numbers of publications in each category included in our review.