Fig. 1.
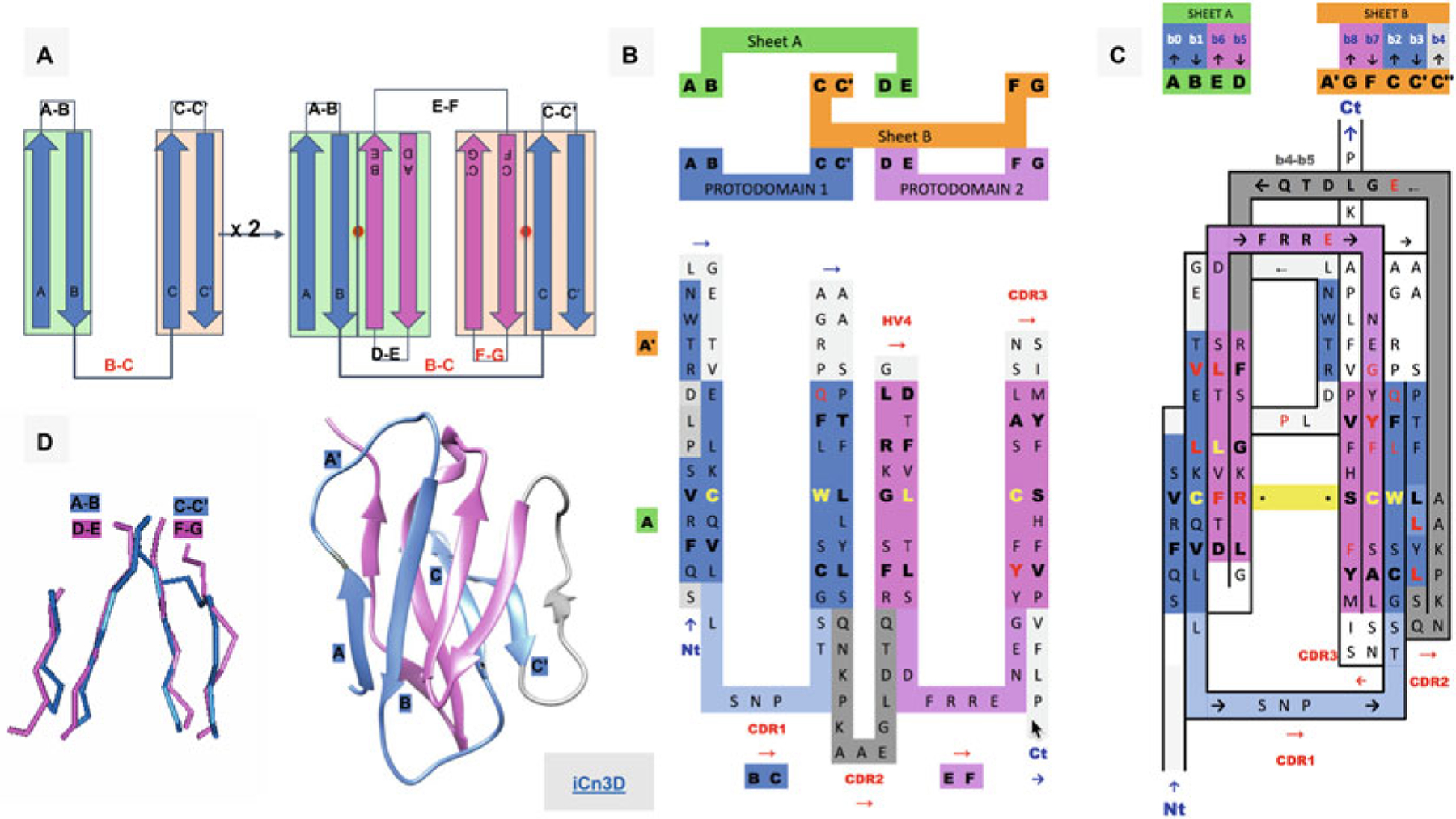
Ig Greek key protodomain topology, duplication, and symmetric arrangement of protodomains. (a) Idealized Ig protodomain motif topology bb-bb, with 2 beta hairpins connected by a Greek key linker (b, c), duplication, and schematic arrangement: A|B + C|C′ = D|E + F|G. Each hairpin theoretically forms to a plane.Each Ig type will present departures from idealized protodomains in their domain context, due to either the protodomain-protodomain linker (not shown here) or some partial structural rearrangement of strand A (see (d) and Fig. 2 for variants in IgV, VNAR, and IgC). Protodomain strands will be displayed blue and magenta for consecutive protodomains 1 and 2, respectively. Planes/Sheets A and B will be consistently shown in green and orange background color. (b) Topology/sequence of consecutive protodomains A|B − C|C′ + D|E − F|G. Interestingly, the well-known CDR1 loop in immunoglobulins appears as the Greek key linker between strands B and C, while the CDR2 is formed by linking the two protodomains (as we shall see in Fig. 2 this is where most Ig domains vary depending of the length and shape of this linker, which presents some secondary structure in the case of IgV giving rise to CDR2). (c) 2D Topology/sequence map strand arrangement of protodomains corresponding to the 3D domain C2 symmetry with the formation of symmetry equivalent B< = >E and C< = >F strand-strand protodomain interface, bringing hairpins A|B and D|E in the same sheet (Sheet A or A|B||E|D) and correlatively bringing hairpins C|C′ and F|G in the same sheet (Sheet B or G|F||C|C′) facing each other as in a sandwich. A simple 3D rotation through a common axis gives a structural correspondence of the two protodomains A|B − C|C′ and D|E − F|G, with a structural alignment (see (d)) varying usually between 1 and 2A in the most distorted cases. The well-known CCW(L) pattern highlighted in yellow is mapped at the protodomain level in symmetrically equivalent positions (see also Fig. 3d). (d) 3D protodomain alignment for a CD8a domain (1CD8) that superimpose with an RMSD of 1.98 (see Fig. 4 for corresponding sequence alignment) showing only structurally aligned residues, with ribbon picture (produced by Chimera [90]) showing strand definitions. Protodomain 1 in blue and protodomain 2 in magenta. Domain visualization with Sheet A in front in the order A|B||E|D’. Link to iCn3D https://d55qc.app.goo.gl/bmCQRj7DWcmqsmna6
