Fig. 6.
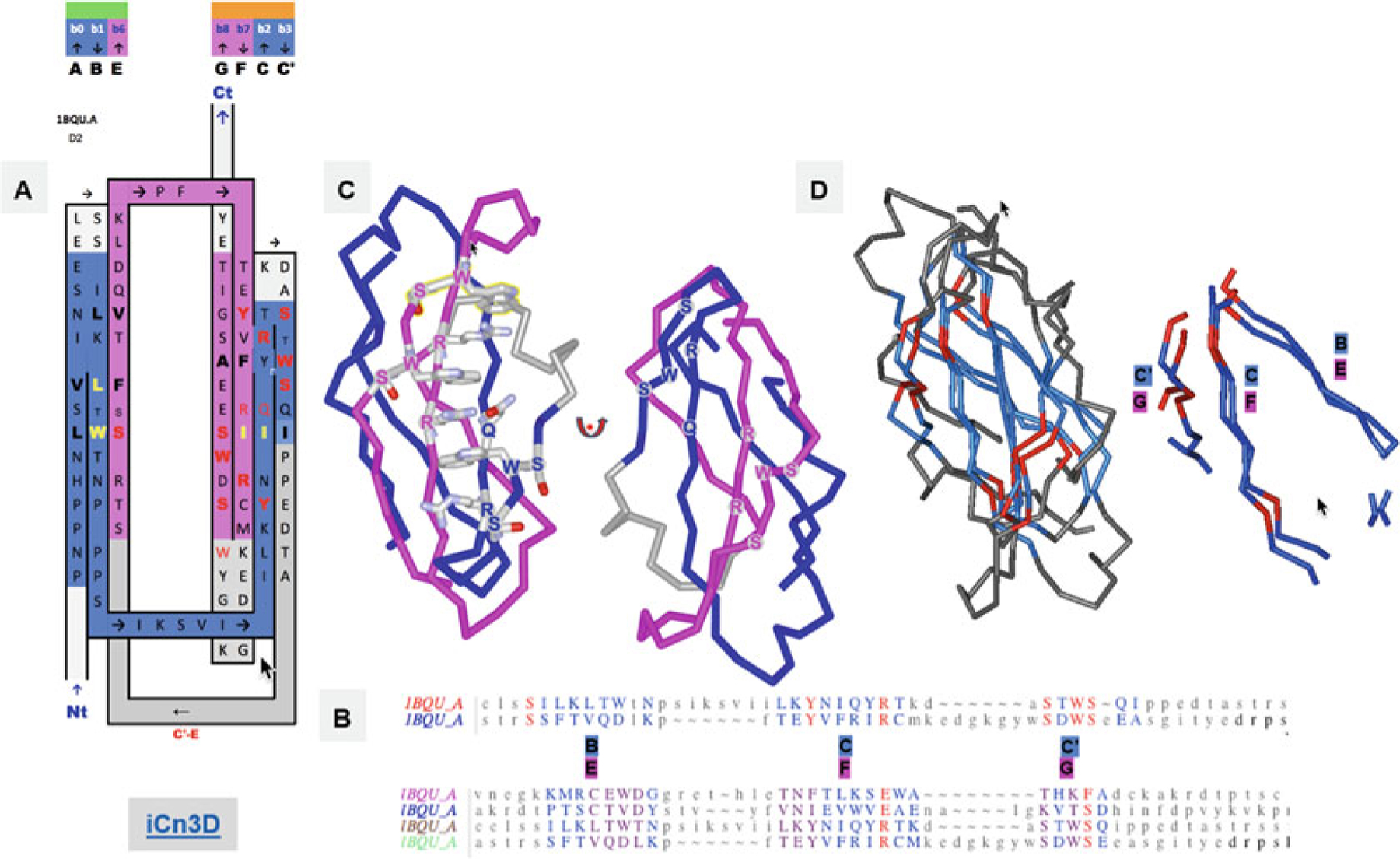
FN3 Ig domains. (a) Another Ig-fold variant, the FN3 superfamily, with the example of the cytokine-binding homology region (CHR) of the cell surface receptor gp130, the second FN3 domain proximal to the membrane surface. The inter-protodomain linker now connects C′–E through a Greek key loop bridging the two sheets, composed of A|B||E and G|F||C|C′. In this case, what would otherwise be a D strand in linking back to the C′ strand (Fig. 2), removing one strand from the other Sheet A (A|B||E(D)) rather than Sheet B (G|F||C(|C′)) as in IgC (see Figs. 2 and 3). The sequence patterns SxWS in strands C′ and G and R/QxR in strands C and F match symmetrically. (b) Structure-based protodomain sequence alignment for domain 2, followed by domain 1 and 2 together, respectively, where one can observe each domain idiosyncratic protodomain “internal conservation” sequence patterns (see text for details). In domain 2 residues S, Y, and R, SxWS are matched, while in domain 1, the pattern is totally different with residues C, D, N, and E. Only one residue S is common to three out of four protodomains, while a R vs. E in symmetrically equivalent strands C and F is observed consistently, a residue which is part of that C||F zipper (see text and Fig. S2). RMSD is 1.8A between domain 2 protodomains and 2.89A for domain 1, 2.2, and 2.5, respectively, vs. domain 2 protodomains (multiple alignment). (c) The symmetric sequence patterns matched in structure forming a cation-pi ladder W*R*W*R*W from both (W)SxWS, the so called WS motif (see text). (d) Structure alignment of the two protodomains matching Strands B-C|C′ and E-F|G that combine as (A|)B||E (Sheet A) and G|F||C|C′ on Sheet B, corresponding to the pairwise protodomain alignment of domain 2 (see sequence alignment in B) https://d55qc.app.goo.gl/DcmpiJy2CVmxtHKN2
