Fig. 7.
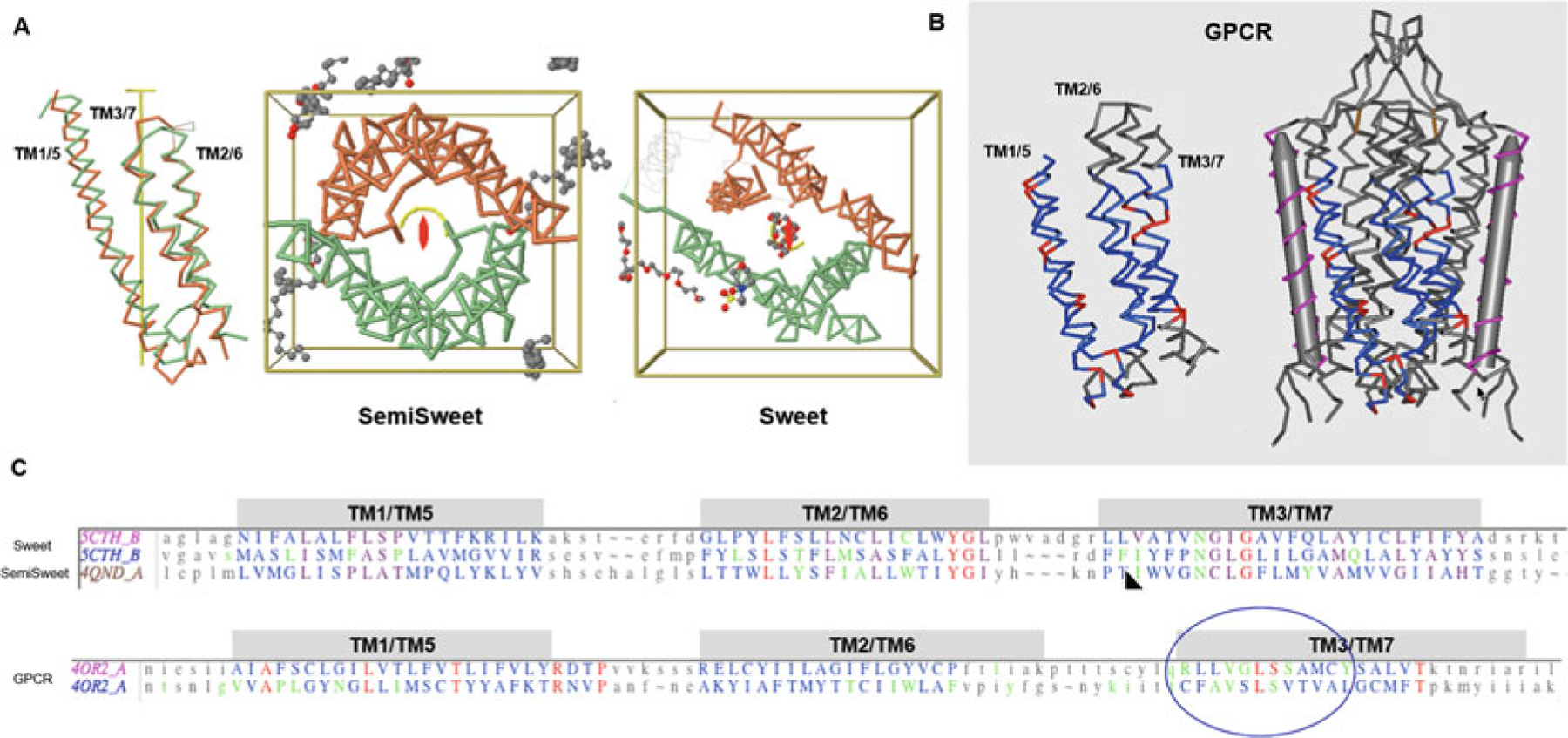
7-Transmembrane helical (7-TMH) proteins. Sweets and GPCRs. (a) Sweet protodomains (3-TMH) aligned, bacterial SemiSweet (3TMH) dimer, and 7-TMH Sweet Protein. The linker between the two Sweet protodomains forms an additional transmembrane helix (TM4). While formed with three consecutive helices in sequence, a protodomain exhibits a 1–3–2 structural arrangement in 3D that is duplicated to form a symmetric pseudosymmetric domain equivalent to a Bacterial SemiSweet symmetric dimer (less TM4). The two protodomains match each other with a RMSD of 1.36 (Sweet protein structure 5CTH) after optimization (automatic detection alignment was 2.91A). A bacterial SemiSweet 3-TMH “domain” aligns with Sweet protodomains with an RMSD of 1.98A (SemiSweet structures 3QND/3QNC). The 7-TMH and 3-TMH dimer align very well not only at the protodomain level but at the dimer vs. pseudo-dimer level. Here displayed with the symmetry axis perpendicular to the plane. The ligand lies on the axis of symmetry. (b) 7-TMH Class C GPCR (structure 4OR2—metabotropic glutamate receptors (mGlus) bound to an allosteric modulator) protodomain optimum alignment with an RMSD of 3.32A through interactive alignment software Cn3D. GPCRs can also be considered with a two-protodomain arrangements. The two protodomains exhibit a distinct 1–2–3 organization in 3D. Here we display the alignment of the whole 7-TMH protein onto itself; the symmetry match can be observed with a solid gray cylinder for the TM4 “linker.” Unlike Sweet, symmetry detection programs do not detect pseudosymmetry systematically but can, in a few cases, using stringent criteria. Interactive alignment of 3-TMH protodomains was used as the method of choice in this case. In all known Class A structures, we have examined, but also in the two Class C structures currently available, pseudosymmetry highlights symmetry equivalent residues in TM1/5, TM2/6, and TM3/7, in a systematic way for some key residues. The structurally aligned TM3/7 helices often exhibit a pseudosymmetric sequence motif (see C and text), framing ligand-binding residues pseudosymmetrically, with ligands lying for a significant part on the axis of symmetry. (c) Associated sequence alignments with mapped ligand-binding residues (or rather residues within a 4A radius from ligand) for Sweet/SemiSweet (sugar) and for the GPCR vs. its ligand. It is important to note that for any pseudosymmetric domain, a protodomain defines a domain entirely (see text). A protodomain is usually idiosyncratic. Here the Sweet protodomain is very different in topology 1-3-2 vs. GPCRs with 1-2-3 topology, yet each defines its domain through that same duplication and pseudosymmetric arrangement
