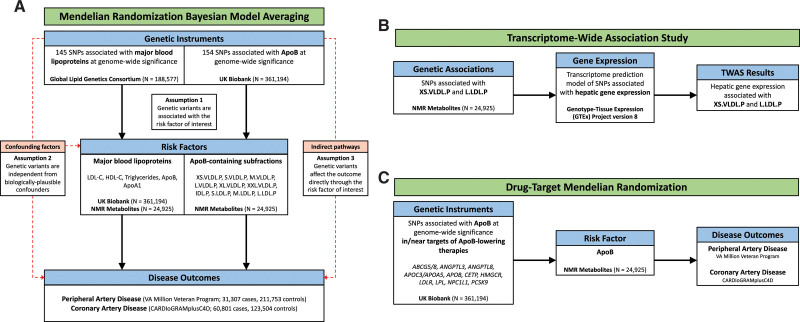
Figure 1.
Overview of risk factor prioritization, drug target, and transcriptome-wide association study (TWAS) analyses. Overview of main analyses. A, Risk factor prioritization was performed with mendelian randomization (MR) bayesian model averaging to prioritize the contribution of major lipoproteins and apolipoprotein (Apo) B–containing subfractions to peripheral artery disease (PAD) and coronary artery disease (CAD) risk. The primary MR assumptions are denoted, with red dashed lines representing violations of the MR assumptions. B, A TWAS integrating gene expression and genetic association data was performed to identify putative genes involved in the regulation of the prioritized ApoB-containing subfractions. C, Drug target MR was performed to identify the effect of genes encoding targets of ApoB-lowering medications on PAD and CAD outcomes. FDR indicates false discovery rate; HDL-C, high-density lipoprotein cholesterol; IDL.P, intermediate-density lipoprotein particles; L.LDL.P, large large-density lipoprotein particles; L.VLDL.P, large very-large-density lipoprotein particles; M.LDL.P, medium large-density lipoprotein particles; M.VLDL.P, medium very-large-density lipoprotein particles; NMR, nuclear magnetic resonance; PAD, peripheral artery disease; S.LDL.P, small large-density lipoprotein particles; SNP, single nucleotide polymorphism; S.VLDL.P, small very-large-density lipoprotein particles; XL.VLDL.P, extralarge very-large-density lipoprotein particles; XS.VLDL.P, extra-small very-large-density lipoprotein particles; and XXL.VLDL.P, extra-extralarge very-large-density lipoprotein particles.