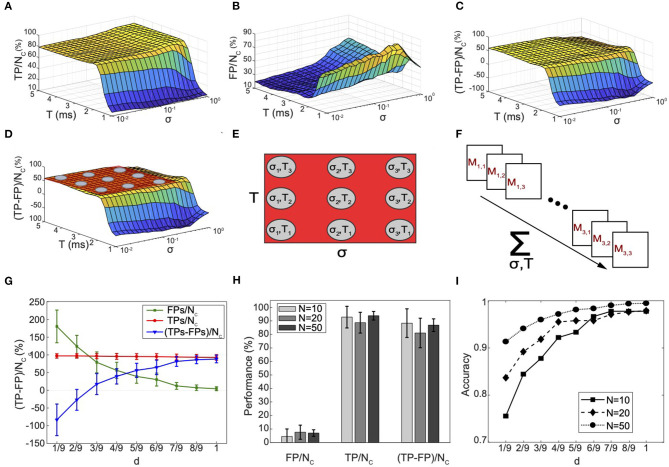
Figure 4.
Connectivity reconstruction and performance evaluation in synthetic neuronal networks. (A–C) Evaluation of the performance of the connectivity method under varying time window (−T, +T), standard deviation σ, and for a fixed ϵ = 0.7 ms, for one simulated network of 10 neurons. (A) Net number of detected true positives TP/nc. (B) Net number of FPs FP/nc. (C) Ratio Δ = (TP−FP)/nc as chosen metric for evaluation. nc is the known total number of connections in the simulated network. (D) Visual highlight on the behavior of Δ: a peak of performance is reached around 65%; low variability is proven for a wide range of T and σ (plateau indicated by the red plane). (E) Definition of a collection of K = 9 points p in the T-σ space for which the algorithm shows performances falling in correspondence of the plateau area. (F) Abstract representation of the statistical method for recognition of FPs connections and connectivity reconstruction. For each point a connectivity matrix Mp is computed, resulting in the computation of K = 9 different connectivity matrices for the same input network. Combination of the results enables computation of the frequency fjk for each connection following Equation (9). (G) Average (N = 20) performances computed at different discrimination threshold d. The TPs remain roughly constant. On the other hand, the FPs decrease and fluctuate around very small percentage. The algorithm filters out the FPs that fluctuate at high frequency reaching best performances (85%) at d = 1. (H) Analysis of scaling properties. An average (N = 20) number of TP/nc, FP/nc, and Δ was computed for 20 randomly generated networks with 10, 20, and 50 neurons, respectively. Very good performances are maintained constant for increasing network size. (I) Accuracy ACC indicator computed for different network sizes as a function of the discrimination threshold d. The error bars stand for the standard deviation of a dataset of 20 different networks.