Fig. 1. FAN1 contains a MIP box that is negatively regulated by CDK-mediated phosphorylation at S126.
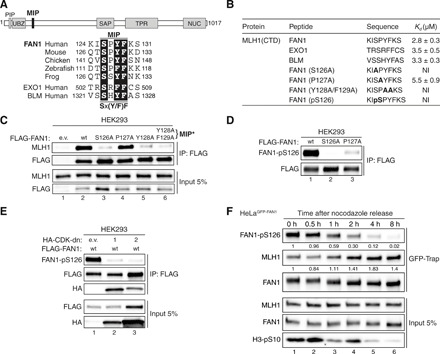
(A) Top: Protein domain architecture of human FAN1 indicating the location of the MIP box. PIP, proliferating cell nuclear antigen–interacting peptide box; UBZ, ubiquitin-binding zinc finger; SAP, SAF-A/B, Acinus, and PIAS; TPR, tetratricopeptide repeat; NUC, nuclease. Bottom: Sequence alignment of the FAN1 orthologs with the MIP box of human EXO1 and BLM. (B) Dissociation constants (Kd) indicating the binding affinity of MLH1 CTD for the indicated peptides from FAN1, EXO1, and BLM as determined by ITC. NI, no interaction (weak and constant signal under the condition used). (C) Immunoblots of FLAG-M2 affinity resin immunoprecipitations (IPs) of extracts from HEK293 cells transfected either with empty vector (e.v.), FLAG-FAN1 wild type (wt), or the variant constructs. The Y128A/F129A mutation is denoted as MIP*. (D) Immunoblots of FLAG-M2 affinity resin IPs of extracts from HEK293 cells transfected with the indicated FLAG-FAN1 constructs. (E) Immunoblots of FLAG-M2 affinity resin IPs of extracts from HEK293 cells cotransfected with FLAG-FAN1 and vectors expressing hemagglutinin (HA)–tagged dominant-negative (dn) forms of CDK1 or CDK2. (F) Immunoblots of green fluorescent protein (GFP)–Trap IPs of extracts from HeLaGFP-FAN1 cells synchronized in mitosis by thymidine-nocodazole block. Mitotic cells were replated in fresh medium for the indicated time periods. The relative ratios of pS126 to FAN1 and MLH1 to FAN1 in GFP-Trap samples were quantified by densitometry using ImageJ. (C to F) The antibodies used are shown on the left.
