Fig. 6. FAN1-MLH1 interaction affects the repair of ICLs and slipped-DNA repeats.
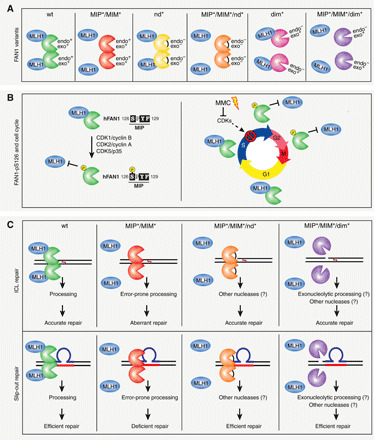
Summary of the FAN1-MLH1 complex interaction and a plausible model for its involvement in ICL and slip-out repair. (A) Scheme of the FAN1-MLH1 interaction in different states of wt or mutated FAN1. Protein orientation is unknown and arbitrarily presented for ease. (B) Left: FAN1 is subjected to CDK-mediated phosphorylation at S126 located within the MIP box. Right: Regulation of FAN1-MLH1 interaction through the cell cycle and in response to ICL damage. (C) Top: The FAN1-MLH1 complex localizes to ICL damaged chromatin to preserve genome stability and ensure cell viability. In cells, devoid of the FAN1-MLH1 interaction aberrant ICL repair and cell death ensues. Inactivation of FAN1’s endo- and exonuclease activities by mutating D960A or inhibition of FAN1 dimerization, affecting FAN1’s endo- but not exonucleolytic activity, would alleviate the toxicity of the FAN1 MIP/MIM-mutated variant and promote cell proliferation and survival. Bottom: Inhibition of the FAN1-MLH1 interaction (MIP*/MIM*) blocks repair of slipped-DNAs. Repair defects were rescued when the MLH1-interacting–defective FAN1 was also defective either in both its endonuclease activities or in dimer formation (endo- but not exonuclease defective). Therefore, the regulatory aspects of the FAN1 binding to MLH1 aligns the pathway of ICL repair with that of slipped-DNA processing. Mutations: MIP*, Y128A/F129A; MIM*, L155A/L159A; nd*, D960A; dim*, K525E/R526E/K528E.
