Abstract
Objective
Glycemic control is an important component of critical care. We present a data-driven method for predicting intensive care unit (ICU) patient response to glycemic control protocols while accounting for patient heterogeneity and variations in care.
Materials and Methods
Using electronic medical records (EMRs) of 18 961 ICU admissions from the MIMIC-III dataset, including 318 574 blood glucose measurements, we train and validate a gradient boosted tree machine learning (ML) algorithm to forecast patient blood glucose and a 95% prediction interval at 2-hour intervals. The model uses as inputs irregular multivariate time series data relating to recent in-patient medical history and glycemic control, including previous blood glucose, nutrition, and insulin dosing.
Results
Our forecasting model using routinely collected EMRs achieves performance comparable to previous models developed in planned research studies using continuous blood glucose monitoring. Model error, expressed as mean absolute percentage error is 16.5%–16.8%, with Clarke error grid analysis demonstrating that 97% of predictions would be clinically acceptable. The 95% prediction intervals achieve near intended coverage at 93%–94%.
Discussion
ML algorithms built on observational data sources, such as EMRs, present a promising approach for personalization and automation of glycemic control in critical care. Future research may benefit from applying a combination of methodologies and data sources to develop robust methodologies that account for the variations seen in ICU patients and difficultly in detecting the extremes of observed blood glucose values.
Conclusion
We demonstrate that EMRs can be used to train ML algorithms that may be suitable for incorporation into ICU decision support systems.
Keywords: critical care, blood glucose, prediction, electronic health records, machine learning
INTRODUCTION
Data science is expected to play a major role in enabling personalized medicine in the critical care setting.1–3 Critically ill patients who enter the intensive care unit (ICU) exhibit considerable heterogeneity, with large variation in treatment response among patients with similar diagnoses.4 The widespread adoption of electronic medical record (EMR) systems and the resulting accumulation of large clinical datasets enables the development of data-driven, precision medicine approaches to addressing clinical heterogeneity. By working well with the large size and complexity commonly found in these, data machine learning (ML) has become the analytic tool of choice.5
ML can play a role in tasks important in ICU precision medicine, such as prediction and state estimation,6 subgroup discovery,3 and development of control algorithms.7 Glycemic control, an important component of critical care, may benefit from all of these ML tasks from the development of algorithms which provide individual patient glucose predictions to the detection early risk of hypo- or extreme hyperglycemia, through individualized control algorithms. In this article, we propose a blood glucose forecasting model, learnt directly from EMRs, with the aim that such models could be used for decision support for management of patient blood glucose in the ICU.
Glycemic control in the ICU is complex, with previous studies using observational datasets demonstrating considerable variation in ICU patient blood glucose levels. Stress-related hyperglycemia, defined as blood glucose levels above 180 mg/dL,8 is common on presentation to the ICU with 1 multi-ICU hospital reporting rates of 9% and 28.6% of nondiabetic and diabetic ICU patients over nearly 20 000 ICU admissions.9 Hyperglycemia evolved as an adaptive response to acute trauma (the “fight or flight” response).10 However, in ICU patients, hyperglycemia is a marker of increased morbidity and mortality11 and is potentially harmful.12 As such, glycemic control is considered a core aspect of critical care,8,13 with insulin therapy the primary approach to controlling blood glucose levels.8,14 However, a further complication is the risk that excessive treatment may cause hypoglycemia, where blood glucose falls below 70 mg/dL, with severe hypoglycemia (< 40 mg/dL) associated with increased mortality.15 Hypoglycemia (< 70 mg/dL) has been reported in 2% and 6% of diabetic and nondiabetic ICU patients on ICU entry9 and 21.3% of ICU patients at least once during their stay.16 As a result, current approaches to glycemic control in the ICU emphasize the avoidance of both hypoglycemia and the extremes of hyperglycemia.
Current guidelines on ICU glucose control were developed following a series of randomized controlled trials between 2001 and 2009 that investigated tight glucose control in the ICU.17–22 Following evidence of an association between average blood glucose in the range near 100 mg/dL and a reduction in ICU mortality, early trials examining efforts to maintain glucose close to this range (within ∼10 mg/dL, referred to as tight glycemic control [TGG]) found positive results.18 However, these were followed by further trials and ultimately the large multicentre NICE-SUGAR trial that reported increased hypoglycemia and mortality in the TGG arm.17 The current clinical consensus is to aim for more moderate control of < 180 mg/dL, with disagreement on the lower bound.8,14 However, while NICE-SUGAR clearly demonstrated that TGG in general is not best practice, there remain open research questions—in particular around the role technology and algorithms can play in glycemic control.23,24
While a broad consensus has been reached that moderate glycemic control should be the clinical standard, there is less evidence on how to achieve and maintain control. Analysis of observational datasets suggests evidence of variation in practice even within single hospitals.9 A wide range of protocols and instruments used to achieve control have been described in the literature, with clear evidence of variation in effectiveness.25 There is an emphasis on using insulin to achieve target blood glucose, with many protocols not accounting for patient feeding,25 despite enteral feeding being frequently interrupted in the ICU due to treatments.26 Indeed, unplanned feeding reduction or interruption may lead to hypoglycemia events.27,28 Further, there is evidence that variation in blood glucose level (or associated metabolic states), independent of the target value, is associated with increased mortality.29,30 This suggests that the development of data-driven approaches that can naturally adapt to variations between and within ICU patients may augment or outperform fixed protocols.
Previous work on developing algorithms for prediction of blood glucose in ICU have largely focused on planned research studies with frequent measures of blood glucose for a small number of subjects rather than utilizing existing records for greater numbers of subjects with much sparser measurements. A large body of literature exists adapting physiology-based systems of ordinary differential equations to the ICU settings.24,31–34 These models generally flow between several idealized compartments such as blood glucose, blood insulin, and interstitial insulin. While this approach may enable accurate predictions, it is difficult to account for additional covariates that are outside the model, and the models generally contain large numbers of parameters that are not identifiable from a typical observational dataset, limiting their transferability between different ICU subpopulations. The incorporation of clinical events, for example administration of vasopressors, is important as these may predict aspects of glucose response in the ICU.12 Data-driven approaches to date have also been planned, such as prospective studies with a large number of measurements on small numbers of patients.35–38 ML algorithms developed using EMRs offer an alternative, or combined approach, to planned studies, which can potentially uncover new aspects important to glycemic control in the ICU because they can readily account for a large number of input features.
We present a purely data-driven method for predicting ICU patient response to glycemic control protocols using an observational EMR dataset. Such models may enable personalization and automation of glycemic control in critical care. We use Catboost, given recent evidence of its gradient boosting outperforming other ML algorithms at forecasting in complex hierarchical time-series datasets and being relatively quick to train.39,40 Additionally, in order to investigate the presence of patient heterogeneity and its impact on predictive performance, we cluster the observed glucose trajectories into subgroups and evaluate the ML algorithms on these groups.
MATERIALS AND METHODS
Data
Our dataset is constructed from the publicly available MIMIC-III database (ethics approval was not required).41 There is variation in the recording of insulin inputs between the EMR applications (MetaVision and CareVue) used to construct MIMIC-III. In the CareVue portion, it is difficult to determine exactly when insulin was administered for some patients. As a result, the CareVue portion of the 60 000 ICU admissions available in MIMIC-III were excluded. The resulting dataset comprises deidentified health data associated with 18 691 ICU stays and 14 742 critical care patients at Beth Israel Deaconess Medical Center in Boston, Massachusetts between 2008 and 2012.
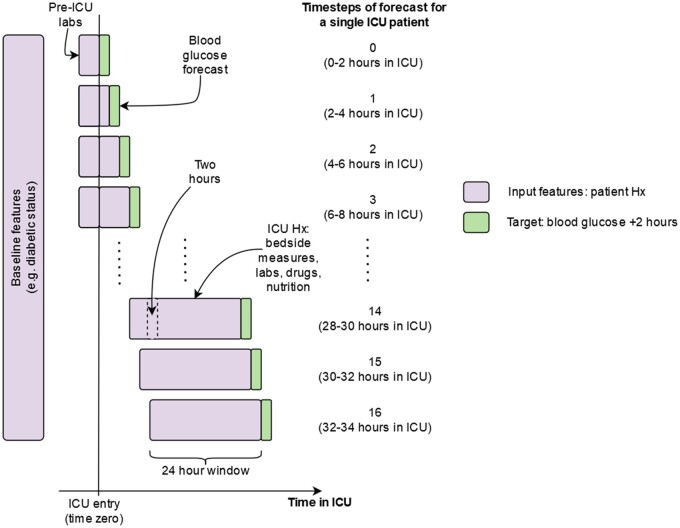
The outcome of interest is the 2-hour ahead point of care blood glucose at each 2-hour time-step from a patient’s entry to ICU (Figure 1). We predict the value and a 95% prediction interval, a range of values in which we expect the observed value to fall 95% of the time. The dataset includes a range of available input features, fully listed in Supplementary Appendix A, including common point of care physiological measures, laboratory results, mechanical ventilation parameters, vasopressors, patient type, amount of dextrose in IV infusions, enteral nutrition, parenteral nutrition and past values of point of care blood glucose, laboratory glucose, and insulin inputs.
Figure 1.
Illustration of the model forecasting process. At each 2-hour time-step from a patient’s entry into ICU we forecast ahead 2 hours. The input features are updated to account for up to 24-hour window of previous values for blood glucose, insulin, and nutritional intake features. For lab and bedside physiological features, the most recently available measure is used. Abbreviations: Hx, history. Given that during the period of analysis blood glucose was typically measured by clinicians at Beth Israel Deaconess on a 2- to 4-hourly schedule, we discretize the dataset to have a time-step every 2 hours.9 Time zero is a patient’s entry to ICU, with any applicable laboratory results carried forward to time zero (see below).
Insulin is recorded as intravenous (IV) infusion or push and bolus injection, with short-, medium-, and long-acting variants available for bolus injections and long and short for IV infusions and pushes. Insulin is coded as the total amount over the 2-hour time intervals for each of the 5 available route/type combinations. Aspects of the dataset construction, in particular the type of insulin inputs, are based on code made available by previous researchers.9,41 Nutritional composition (total calories and percentage of carbohydrates, sugar, fat, and protein) of enteral nutritional products come from the manufacturers’ websites (full details listed in Supplementary Appendix B), while parenteral nutrition composition is as recorded in the dataset or imputed based on average composition.
Missing data and irregular measurement frequencies are a common issue in analysis of EMRs.42 In order to deal with irregular measurements, we carry forward the last value of each input feature and calculate the elapsed time since that measurement. Both the value of the input feature (possibly carried forward) and the elapsed time are used as input features in model development. For blood glucose, insulin, and nutritional intake features, we additionally carry forward the previous 24 hours’ worth of values as new input features in order to account for potential delays in action and correlations over time (Figure 1).
Models
We develop 2 ML approaches using the Catboost gradient boosting library.39 These models were chosen as they present alternative approaches to predicting both a point estimate and uncertainty quantification through probabilistic forecasting. The first is a Catboost regression model with dual estimation of the expected outcome and the standard deviation of the prediction distribution, the ‘uncertainty regression’ model.43 This form of estimation can be performed using the class CatBoostRegressor with the argument loss_function=“RMSEWithUncertainty” in the Python version of Catboost 2.4. The second model is a combination of quantile regressions with models for quantiles of 0.025, 0.5, and 0.975, the “quantile regression” model. This form of estimation can be performed similar to the above, for example for the 0.975 quantile as CatBoostRegressor (loss_function=“Quantile: alpha = 0.95”, …). For both the quantile and uncertainty model the glucose is modeled as the (natural) logarithm transformed blood glucose, resulting in an approximately normally distributed variable. Calculation of the Box-Cox transformation exponent suggests an optimal transform of −0.3, with the logarithm (corresponding to 0.0) being chosen to simplify back-transformation.44 The point estimate (mean value) for blood glucose is calculated as where and are the estimates of the mean and standard deviation on the logarithmic scale. The 95% prediction interval is calculated as . For the quantile model, the point estimate is given by where , with and corresponding to estimates of the 2.5th and 97.5th percentiles of the logarithm transformed glucose. The 95% prediction interval is calculated as . Additionally, the performance of algorithms is benchmarked against using the previous blood glucose measure as the forecast (last observation carried forward).
We perform analyses using R version 3.545 and Python version 3.7, prepare data using data.table version 1.18,46 produce graphics using ggplot2 version 3.3,47 and fit the models using Catboost version 2.4. Code for the analysis can be found at https://github.com/oizin/glucose-data-driven-prediction.
Model validation
The dataset is randomly split into a 70% training (13 279 ICU admissions) and 30% test (5682 ICU admissions) sets. Sample splits are performed by ICU admission ID to avoid potential information leakage. We evaluate all models on the test set only after finalization of hyperparameter settings to ensure unbiased assessments of model generalizability. As the algorithms were computationally expensive to train, we perform hyperparameter tuning by randomly splitting the training set into 80% development and 20% validation sets.
Performance metrics
We calculate several model performance metrics to assess the ability of the model to accurately provide point estimates in clinically meaningful circumstances, and to provide adequate indications of prediction uncertainty. Clarke error grid analysis (CEGA) was designed to compare a new blood glucose measurement method with a reference standard.48 In this case, the model prediction corresponds to the new approach, with the reference value—point of care blood glucose measurement. CEGA divides predictions into 5 groups that are classified based on A) predictions within 20% of the reference value, B) predictions that are outside of 20% but would not lead to inappropriate treatment, C) predictions leading to unnecessary treatment, D) predictions that fail to identify a potentially dangerous hypoglycemic or hyperglycemic event, and E) predictions that would confuse hypoglycemia for hyperglycemia and vice versa. We additionally report the root mean square error (RMSE), the mean absolute percentage error (MAPE), median absolute deviation (MAD), and the coverage of the 95% prediction intervals across the entire sample and for specific blood glucose ranges. The RMSE and MAD both quantify the average deviation of a prediction from the measured value. However, the RMSE places relatively greater weight on larger deviations compared to the MAD. The MAPE is the MAD divided by the measured value, it is the “percentage error.” Prediction interval coverage is calculated as the proportion of time the observed blood glucose value falls inside the prediction interval.
Performance by cluster
To assess the performance of the models across patient subgroups, we cluster the ICU admission blood glucose trajectories using the CLARA clustering algorithm.49 This algorithm is designed to scale on large datasets by clustering on subsamples and then aggregating the results across the whole dataset. The Euclidean distance is chosen as the measure of similarity between patients. The number of clusters is determined via the elbow method as the point of inflection in the curve describing the decrease within cluster variation with the number of clusters. We report the patient daily average blood glucose level for day 1 and 2 of their ICU stay, diabetic status, and 30-day mortality rate for each cluster.
Model interpretation
For each feature we calculate the associated change in the loss function that would occur with removal of the feature to assess its importance in forecasting blood glucose.39
RESULTS
Cohort description
Our dataset includes 14 742 patients. Accounting for readmission, the total number of ICU admissions is 18 691 with an average stay of 3.9 days. Most admissions are nondiabetic (76.7%), and the cohort is evenly split between medical (42%) and surgical (46%) ICU patients, with a small number of cardiac ICU admissions (11%). There are 318 574 glucose measurement events, an average of 4.3 per patient per day with 22.5% of these above 170 mg/dL and 1.3% below 70 mg/dL. Insulin is administered fairly frequently during ICU admissions (37.0% of admissions), with the most common route of insulin administration being bolus injection (31.3% of admissions), followed by IV infusion (19.9% of admissions). Nutritional intake information, in the form of enteral and parenteral nutrition, is present for 18.0% and 3.5% of admissions. Further details on the cohort are available in Table 1 including a comparison of the training and test splits. As with all EMR systems, absence of a record cannot be taken as absence of an event.41
Table 1.
Descriptive statistics of the analysis cohort
| Description | Overall | Training | Test |
|---|---|---|---|
| ICU admission characteristics | |||
| Number of ICU admissions | 18 961 | 13 279 | 5682 |
| Number of patients | 14 742 | 10 938 | 5172 |
| Age (median) | 66 | 66 | 65 |
| Female (%) | 43.5 | 56.5 | 56.6 |
| Male (%) | 56.5 | 43.5 | 43.4 |
| Type of ICU admission | |||
| Medical | 8026 (42%) | 5607 (42%) | 2419 |
| Surgical | 8787 (46%) | 6170 (46%) | 2617 |
| Cardiac | 2148 (11%) | 1502 (11%) | 646 |
| Type of admission | |||
| Elective | 2774 (14.6%) | 1996 (15.0%) | 778 (13.7%) |
| Emergency/urgent | 16 187 (85.0%) | 11 283 (85.0%) | 4904 (86.3%) |
| Diabetic status | |||
| Diabetic (%) | 23.3 | 23.4 | 23.1 |
| Nondiabetic (%) | 76.7 | 76.6 | 76.9 |
| Glycemic control | |||
| Number of serum blood glucose measurements | 318 574 | 224 315 | 94 259 |
| Above 170 mg/dL (%) | 71 565 (22.5%) | 50 429 | 21 136 |
| Below 70 mg/dL (%) | 4054 (1.3%) | 2761 | 1293 |
| Median number of serum blood glucose measurements per ICU admission (25th, 75th percentiles) | 10 (5, 19) | 10 (5, 19) | 10 (5, 19) |
| Average blood glucose on day of ICU admission | 139.1 mg/dL | 138.9 mg/dL | 141.7 mg/dL |
| Number above 170 mg/dL (%) | 3518 (18.6%) | 2427 (18.3%) | 1091 (19.2%) |
| Number below 70 mg/dL (%) | 55 (0.3%) | 32 (0.2%) | 23 (0.4%) |
| ICU admissions with insulin administered during stay | 7030 (37.1%) | 4982 (37.5%) | 2048 (36.0%) |
| Infusion (average hours) | 3775 (26.8) | 2688 (26.4) | 1067 (28.1) |
| Bolus injection (average times) | 5952 (2.5) | 4224 (2.5) | 1728 (2.6) |
| IV push (average times) | 2583 (2.6) | 1882 (2.6) | 701 (2.7) |
| Nutrition | |||
| ICU admissions with enteral nutrition | 3398 (18.0%) | 2405 (18.1%) | 993 (17.5%) |
| ICU admissions with parenteral nutrition | 655 (3.5%) | 470 (3.5%) | 185 (3.3%) |
| Other ICU admissions and treatments | |||
| ICU admissions with mechanical ventilation | 8868 (46.8%) | 6247 (47.0%) | 2621 (46.1%) |
| ICU admissions with vasopressors administered | 3449 (18.2%) | 2432 (18.3%) | 1017 (17.9%) |
| ICU stay outcomes | |||
| Average length of ICU stay | 3.9 days | 3.9 days | 3.9 days |
| Died within 30 days of ICU stay | 14.4% | 14.4% | 14.5% |
Model performance
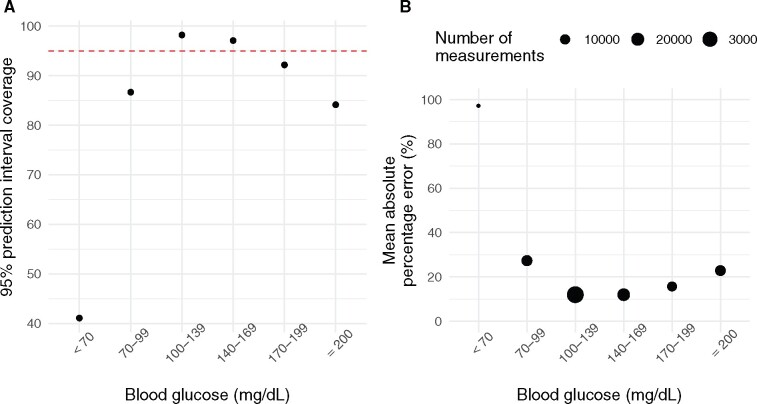
The overall performance of the models is outlined in Table 2. The 95% prediction intervals for the quantile regression and uncertainty regression models include 94% and 93%, respectively, of the measured postadmission blood glucose values. There was no clear difference between the models in terms of point prediction error (MAPE and RMSE), with the quantile regression model marginally better. Both models perform best in the most frequently observed glycemic range (100–200 mg/dL), followed by the hyperglycemic (≥ 200 mg/dL) and then hypoglycemic ranges (< 70 mg/dL), with the uncertainty regression model achieving MAPEs of 12.4%, 22.5%, and 96.3% for these ranges. Additionally, there is variation in model performance by ICU admission type, with the uncertainty regression model performance highest for surgical (including cardiac surgery) (MAPE: 15.1%), followed by cardiac (MAPE: 17.9%) and medical admissions (MAPE: 19.0%). We show further breakdown of the results below (Figures 1–3) for the uncertainty regression model. These illustrate that the majority (97.1%) of model predictions lead to clinically acceptable decisions (Figure 2) and that the performance of the model varies by observed blood glucose value with better performance in terms of point prediction accuracy and prediction interval coverage for blood glucose values in the range 100–200 mg/dL (Figure 3). We see that due to the difficulty of predicting hypoglycemic, and to a lesser extent, hyperglycemic events, the prediction intervals are conservative in the range 100–169 mg/dL near and below intended coverage (95%) elsewhere. We provide additional model performance metrics stratified by factors such as admission diagnosis, admission type, and temporal factors in Supplementary Appendix.
Table 2.
Model performance metrics for blood glucose forecasting models using forecast and observed blood glucose values from the test dataset
| Metric | Quantile regression (Catboost) | Uncertainty regression (Catboost) | Last observation carried forward |
|---|---|---|---|
| RMSE (mg/dL) | 49.7 | 49.9 | 65.5 |
| MAPE (%) | 16.5 | 16.8 | 21.2 |
| MAD (mg/dL) | 15.4 | 16.1 | 20.0 |
| Coverage (nominal 95%) | 94.0% | 93.2% | n/a |
Figure 2.
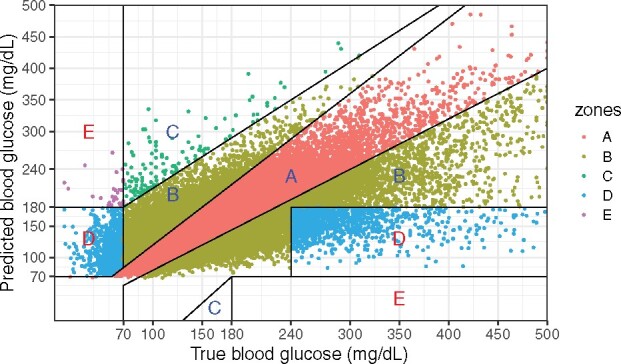
Clarke error grid analysis using forecast and observed blood glucose values from the test dataset. The percentage of points in each region are A: 71.4%; B: 25.6%; C: 0.2%; D:2.8; E:< 0.1, with the regions ranked (A–E) in terms of increasingly dangerous clinical consequences for the prediction error.
Figure 3.
(A) Prediction interval coverage by measured blood glucose value using forecast and observed blood glucose values from the test dataset. The overall coverage is 93.2%. (B) Mean absolute percentage error by measured blood glucose value. We see a pattern of poor performance in prediction of hypoglycemic and, to a lesser extent, hyperglycemic events.
Performance by cluster
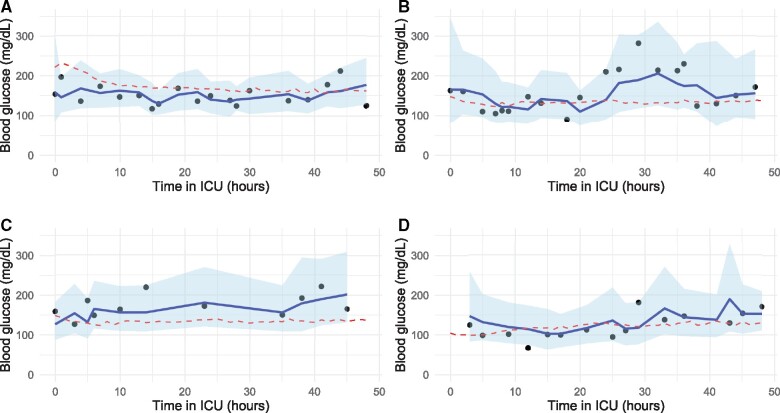
Based on the elbow method the blood glucose trajectories were clustered into 3 clusters. Cluster descriptive statistics and performance metrics are displayed in Table 3. Cluster 1 contained the largest percentage of diabetics and had the highest blood glucose values and 30-day mortality rates, with clusters 2 and 3 exhibiting increasingly fewer diabetic admissions and lower mortality and blood glucose values, respectively. Based on the MAPE and MAD, the model performs best for patients in Cluster 2, those patients with the greatest stability in their blood glucose measurements. Examples of observed blood glucose values, forecasts, and the cluster means for 4 patients are shown in Figure 4.
Table 3.
Cluster descriptive statistics and model performance metrics for the uncertainty regression model
| Cluster 1 | Cluster 2 | Cluster 3 | |
|---|---|---|---|
| Cluster descriptive statistics | |||
| % of admissions | 26.8 | 51.8 | 21.4 |
| Mean (SD) blood glucose | |||
| Day 1 | 200.6 (80.0) | 131.8 (96.0) | 104.5 (30.4) |
| Day 2 | 170.2 (69.5) | 133.6 (42.5) | 120.5 (42.5) |
| Day 3 | 163.5 (64.5) | 133.6 (43.7) | 124.4 (43.1) |
| Diabetic (%) | 44.4 | 22.9 | 11.0 |
| Died within 30 days of ICU stay | 17.8 | 15.5 | 9.4 |
| Model performance metrics | |||
| RMSE (mg/dL) | 47.7 | 58.7 | 30.2 |
| MAPE (%) | 18.6 | 15.4 | 16.7 |
| MAD (mg/dL) | 21.1 | 14.2 | 13.9 |
| Coverage (nominal 95%) | 92.5 | 93.8 | 93.2 |
Figure 4.
Example glucose and prediction trajectories for 4 patients. The model forecasts blood glucose in 2 hours, and these graphs unroll the predictions over a 48-hour period: (A) is a patient from Cluster 1, (B) and (C) are patients from Cluster 2, (D) is a patient from Cluster 3. The red dashed lines indicate the relevant cluster mean trajectories.
Model interpretation
The variable importance measures for the 30 most important features are shown in Table 4.
Table 4.
Feature importance values for the uncertainty regression model
| Feature | Loss function (RMSE) reduction |
|---|---|
| Blood glucose (most recent) | 30.74 |
| Time to blood glucose measurement (most recent) | 6.22 |
| Time of day (hours) | 5.66 |
| Insulin infusion amount over past 2 hours | 4.02 |
| Blood glucose (second most recent) | 3.21 |
| Blood glucose (fourth most recent) | 2.05 |
| Dextrose input in ongoing IV fluid | 1.86 |
| Blood glucose (third most recent) | 1.39 |
| Patient weight | 1.23 |
| Cardiac ICU admission | 1.14 |
| Insulin injection (short-acting) amount over past 10 hours | 1.02 |
| Diabetic patient | 1.00 |
| Insulin injection (short-acting) amount over past 20 hours | 0.91 |
| Insulin injection (short-acting) amount over 2–4 hours ago | 0.83 |
| Insulin injection (short-acting) amount over past 4 hours | 0.81 |
| Insulin injection (short-acting) amount over 4–6 hours ago | 0.81 |
| Blood glucose (fifth most recent) | 0.77 |
| Insulin injection (short-acting) amount over past 4 hours | 0.62 |
| Volume of ongoing enteral nutrition | 0.55 |
| Most recent blood lactate value | 0.55 |
| Insulin injection (short-acting) amount over past 8 hours | 0.54 |
| Insulin injection (short-acting) amount over past 18 hours | 0.48 |
| Insulin infusion amount over past 2 hours | 0.48 |
| Insulin injection (short-acting) amount over past 22 hours | 0.45 |
| Most recent heart rate measurement | 0.44 |
| Time to blood glucose measurement (second most recent) | 0.44 |
| Most recent creatinine result | 0.44 |
| Most recent systolic blood pressure | 0.43 |
| Sugar content in IV fluid over past 4 hours | 0.41 |
| Most recent sodium result | 0.37 |
DISCUSSION
We report a predictive ML algorithm using routinely collected EMR data that takes as input a patient’s medical history during their ICU stay, and at baseline, and forecasts blood glucose at 2-hour ahead time-steps from ICU entry. The model achieves performance that is comparable to previous models developed in planned research studies using continuous blood glucose monitoring devices and, like these, performs better for some patient groups and blood glucose values with relatively poor performance for predicting hypoglycemia.
Our proposed ML model demonstrates a high degree of accuracy in forecasting blood glucose in the range 70–200 mg/dL. The overall and surgical patient MAPEs of 16.5% and 15.1% for 2-hour forecasts of serum blood glucose compares with 15.9% (for a 135-min forecast) achieved in a planned research study on surgical ICU patients using continuous glucose monitoring.35 While other research has achieved lower error margins, this was through shorter forecasting timeframes (30–75 min) reducing comparability.36–38 The current approach aimed to have the potential to assist with glycemic control as practiced in the ICUs studied, with the forecast timeframe a lower bound on the frequency of observed blood glucose measurement.9 As assessed using CEGA, the majority of predictions (97.2%) will lead to a clinically appropriate response. The use of prediction intervals is a strength, with quantification of uncertainty important for glycemic prediction due to potentially complex dynamics decreasing point prediction performance50 and the presence of measurement error.23
One strength of data-driven approaches to glucose prediction in the ICU is the ability to incorporate a wide range of predictor variables, as seen in previous research.35–38 This contrasts with mathematical modeling approaches that generally rely on previous blood glucose measures along with insulin and glucose intake information.24 However, as noted in previous research,35 a risk of ML methods is their “black-box” nature and the potential learning of spurious relationships local to the dataset. Indeed, an approach to comprehensive interrogation (from a physiological standpoint) of the data-driven approaches used in the past research is missing from the literature. In the current research, the variable importance measures are unsurprising, with previous measures of blood glucose, dextrose/carbohydrate intake, diabetes, insulin, and patient weight being of high predictive importance. The presence of ICU unit type as a relatively important feature may account for variation in patient characteristics and treatment patterns unaccounted for in the modeled input features.
Our model performs best in cardiac surgical ICU patients. Generally, these patients are seen as the least critically ill51 and have been a focus of several previous studies using data-driven methods to forecast blood glucose in the ICU.35,36,38,52 However, no previous articles have compared predictive performance between medical and surgical patients for large numbers of patients. The greater stability in their metabolic state may ease forecasting through less shift in their blood glucose distribution over time or with treatment (eg, insulin). Blood glucose in metabolically unstable groups, such as patients with diabetic ketoacidosis (MAPE: 26.5%), or emergency compared to elective admissions in general (MAPE: 17.3% vs 15.0%), is more difficult to predict. Previous theoretical research using dynamic models has shown that time dependent variations in insulin resistance and feeding patterns common to ICU patients can result in complex glucose responses.32,50,53This further highlights the importance of quantifying uncertainty.
EMRs are a messy data source not designed for medical research. However, the current research has highlighted their practical value in the development of predictive models. A major strength of the current ML based approach is the easy incorporation of large numbers of input features which can be modeled without manual feature engineering. Further, the model easily accounted for the irregularly recorded and missing data common in EMRs. With up to 60% of blood glucose measurements not observed at every 2-hour time-step, this was of vital importance. Indeed, the model achieved a level of performance comparable to previous planned research studies based on extensive measurement of a small number of patients. This demonstrates that using sparser data from many patients should be a key aspect of future research on glycemic prediction or control algorithms. The current research has limitations. As noted, performance was worse for more extreme values of blood glucose. In order to have clinical utility, it is important that the model can detect hyperglycemia and hypoglycemia. Detection of hyperglycemia was only slightly worse than values in the ICU normal blood glucose range. However, similar to previous research, our point estimates were unable to detect hypoglycemia at 2-hour forecasts.35 However, by forecasting an interval, we increase the potential to flag circumstances in which hypoglycemia is a risk, with 41% of hypoglycemic events captured within the prediction intervals. A postanalysis investigation of a 2-hour ahead hypoglycemic prediction model showed poor results with the model unable to do more than predict the base rate of ∼1.3% (Supplementary Appendix D). This difficulty of forecasting hypoglycemic clearly warrants further research and analysis. An issue is ensuring the accuracy of a hypoglycemic reading, with the potential that bedside capillary measurements are not an accurate enough tool in critically ill patients.54 Indeed, previous research has identified poor perfusion as a cause of inaccurate hypoglycemia measurements.55 In the current study patients, the average diastolic blood pressure was 56.8 mmHg during hypoglycemic events compared with an average measure of 60.4 mmHg. Alternatively, oscillation in blood glucose, as seen in theoretical ultradian insulin–glucose dynamic model studied in previous research53 may account for transient hypoglycemia.
It is also important to consider the extent to which these results are dataset specific. As the data are from a single institution, the degree to which institutional factors impact the results cannot be quantified. There exists evidence of variation in glycemic control protocols between institutions,26 and future research would be required to investigate the degree of model retraining required to adapt to local protocols. The dataset also constrained the timeframe of predictions. Previous research using continuous blood glucose monitoring devices could be trained on and, hence, predict blood glucose every 5 min for up to 2 hours. Using a sparsely measured EMR to predict over such short time frames would likely be an unjustifiable extrapolation given evidence of nonlinear glucose dynamics.53 Both of these limitations point a way forward for future research. Through pooling EMRs from several institutions, along with data from continuous blood glucose monitoring devices (for potentially a subset of a patients), we may be able to develop robust predictive models.
CONCLUSION
In conclusion, we demonstrate that EMRs can be used to develop blood glucose prediction models that achieve a high degree of accuracy. There is strong potential for developing EMR-based ICU decision support systems for glycemic control and other key ICU physiological parameters. The uncertainty in blood glucose dynamics necessitates the quantification of uncertainty in any forecasts which we demonstrate can be achieved using ML tools. There remain challenges, such as detection of hypoglycemia and assessing the impact of variation of institutional glycemic control policies on model performance. Future research on glycemic control will benefit from greater emphasis on incorporation of real-world data, such as those from EMRs.
FUNDING
This work was supported by the Commonwealth Industrial and Scientific Research Organisation (CSIRO), eHealth NSW, and the Australian government through an Australian Government Research Training Program scholarship and CSIRO top up scholarship.
AUTHOR CONTRIBUTIONS
OF conceived the idea for the study. OF and OPC conceived the initial design of the study. All authors provided critical methodological feedback and contributed to the design of the study. OF prepared the dataset for the study and performed the analysis. OF and LJ drafted the article. All authors contributed to critical revision of the manuscript.
SUPPLEMENTARY MATERIAL
Supplementary material is available at Journal of the American Medical Informatics Association online.
DATA AVAILABILITY STATEMENT
The data underlying this article are available at the MIMIC-III open source database at https://mimic.physionet.org/, and can be accessed following completion of required training as detailed at https://mimic.physionet.org/gettingstarted/access/.
CONFLICT OF INTEREST STATEMENT
None declared.
Supplementary Material
REFERENCES
- 1. Vincent J-L. The coming era of precision medicine for intensive care. Crit Care 2017; 21 (Suppl 3): 314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Maslove DM, Lamontagne F, Marshall JC, Heyland DK.. A path to precision in the ICU. Crit Care 2017; 21 (1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Seymour CW, Gomez H, Chang C-CH, et al. Precision medicine for all? Challenges and opportunities for a precision medicine approach to critical illness. Crit Care 2017; 21 (1): 257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cecconi M, Evans L, Levy M, Rhodes A.. Sepsis and septic shock. Lancet 2018; 392 (10141): 75–87. [DOI] [PubMed] [Google Scholar]
- 5. Yadav P, Steinbach M, Kumar V, Simon G.. Mining electronic health records (EHRs): a survey. ACM Comput Surv 2018; 50 (6): 1–40. [Google Scholar]
- 6. Johnson AE, Ghassemi MM, Nemati S, Niehaus KE, Clifton DA, Clifford GD.. Machine learning and decision support in critical care. Proc IEEE Inst Electr Electron Eng 2016; 104 (2): 444–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Komorowski M, Celi LA, Badawi O, Gordon AC, Faisal AA.. The artificial intelligence clinician learns optimal treatment strategies for sepsis in intensive care. Nat Med 2018; 24 (11): 1716–20. [DOI] [PubMed] [Google Scholar]
- 8. Rhodes A, Evans LE, Alhazzani W, et al. Surviving sepsis campaign: international guidelines for management of sepsis and septic shock: 2016. Intensive Care Med 2017; 43 (3): 304–77. [DOI] [PubMed] [Google Scholar]
- 9. Baker L, Maley JH, Arévalo A, et al. Real-world characterization of blood glucose control and insulin use in the intensive care unit. Sci Rep 2020; 10 (1): 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Van Cromphaut S. Hyperglycaemia as part of the stress response: the underlying mechanisms. Best Pract Res Clin Anaesthesiol 2009; 23 (4): 375–86. [DOI] [PubMed] [Google Scholar]
- 11. Bagshaw SM, Egi M, George C, Bellomo R, Committee ADM.. Early blood glucose control and mortality in critically ill patients in Australia. Crit Care Med 2009; 37 (2): 463–70. [DOI] [PubMed] [Google Scholar]
- 12. Dungan KM, Braithwaite SS, Preiser J-C.. Stress hyperglycaemia. Lancet 2009; 373 (9677): 1798–807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Vincent J-L. Give your patient a fast hug (at least) once a day. Crit Care Med 2005; 33 (6): 1225–9. [DOI] [PubMed] [Google Scholar]
- 14. Qaseem A, Chou R, Humphrey LL, et al. ; for the Clinical Guidelines Committee of the American College of Physicians. Inpatient glycemic control: best practice advice from the Clinical Guidelines Committee of the American College of Physicians. Am J Med Qual 2014; 29 (2): 95–8. [DOI] [PubMed] [Google Scholar]
- 15. Egi M, Bellomo R, Stachowski E, et al. Hypoglycemia and outcome in critically ill patients. Mayo Clin Proc 2010; 85 (3): 217–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cook CB, Kongable GL, Potter DJ, Abad VJ, Leija DE, Anderson M.. Inpatient glucose control: a glycemic survey of 126 US hospitals. J Hosp Med 2009; 4 (9): E7–E14. [DOI] [PubMed] [Google Scholar]
- 17.The NICE-SUGAR Study Investigators. Intensive versus conventional glucose control in critically ill patients. N Engl J Med 2009; 360 (13): 1283–97. [DOI] [PubMed] [Google Scholar]
- 18. Van Den Berghe G, Wouters P, Weekers F, et al. Intensive insulin therapy in critically ill patients. N Engl J Med 2001; 345 (19): 1359–67. [DOI] [PubMed] [Google Scholar]
- 19. Van den Berghe G, Wilmer A, Hermans G, et al. Intensive insulin therapy in the medical ICU. N Engl J Med 2006; 354 (5): 449–61. [DOI] [PubMed] [Google Scholar]
- 20. Arabi YM, Dabbagh OC, Tamim HM, et al. Intensive versus conventional insulin therapy: a randomized controlled trial in medical and surgical critically ill patients. Crit Care Med 2008; 36 (12): 3190–7. [DOI] [PubMed] [Google Scholar]
- 21. Brunkhorst FM, Engel C, Bloos F, et al. Intensive insulin therapy and pentastarch resuscitation in severe sepsis. N Engl J Med 2008; 358 (2): 125–39. [DOI] [PubMed] [Google Scholar]
- 22. De La Rosa GDC, Donado JH, Restrepo AH, et al. Strict glycaemic control in patients hospitalised in a mixed medical and surgical intensive care unit: a randomised clinical trial. Crit Care 2008; 12 (5): R120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Egi M, Finfer S, Bellomo R.. Glycemic control in the ICU. Chest 2011; 140 (1): 212–20. [DOI] [PubMed] [Google Scholar]
- 24. Chase JG, Benyo B, Desaive T.. Glycemic control in the intensive care unit: a control systems perspective. Annu Rev Control 2019; 48: 359–68. [published Online First: Epub Date]. [Google Scholar]
- 25. Wilson M, Weinreb J, Hoo GWS.. Intensive insulin therapy in critical care: a review of 12 protocols. Diabetes Care 2007; 30 (4): 1005–11. [DOI] [PubMed] [Google Scholar]
- 26. McClave SA, Taylor BE, Martindale R, et al. ; the Society of Critical Care Medicine. Guidelines for the provision and assessment of nutrition support therapy in the adult critically ill patient: Society of Critical Care Medicine (SCCM) and American Society for Parenteral and Enteral Nutrition (ASPEN). JPEN J Parenter Enteral Nutr 2016; 40 (2): 159–211. [DOI] [PubMed] [Google Scholar]
- 27. Juneja R, Roudebush CP, Nasraway SA, et al. Computerized intensive insulin dosing can mitigate hypoglycemia and achieve tight glycemic control when glucose measurement is performed frequently and on time. Crit Care 2009; 13 (5): R163– 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Elia M, De S.. A. Tight glucose control in intensive care units: an update with an emphasis on nutritional issues. Curr Opin Clin Nutr Metab Care 2008; 11 (4): 465–70. [DOI] [PubMed] [Google Scholar]
- 29. Krinsley JS. Glycemic control in the critically ill: what have we learned since NICE-SUGAR? Hosp Pract (1995) 2015; 43 (3): 191–7. [DOI] [PubMed] [Google Scholar]
- 30. Uyttendaele V, Dickson JL, Shaw GM, Desaive T, Chase JG.. Untangling glycaemia and mortality in critical care. Crit Care 2017; 21 (1): 152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hovorka R, Chassin LJ, Ellmerer M, Plank J, Wilinska ME.. A simulation model of glucose regulation in the critically ill. Physiol Meas 2008; 29 (8): 959–78. [DOI] [PubMed] [Google Scholar]
- 32. Lin J, Lee D, Chase JG, et al. Stochastic modelling of insulin sensitivity and adaptive glycemic control for critical care. Comput Methods Programs Biomed 2008; 89 (2): 141–52. [DOI] [PubMed] [Google Scholar]
- 33. Chase JG, Shaw GM, Wong X-W, Lotz T, Lin J, Hann CE.. Model-based glycaemic control in critical care—A review of the state of the possible. Biomed Signal Process Control 2006; 1 (1): 3–21. [Google Scholar]
- 34. Dickson JL, Stewart KW, Pretty CG, et al. Generalisability of a virtual trials method for glycaemic control in intensive care. IEEE Trans Biomed Eng 2018; 65 (7): 1543–53. [DOI] [PubMed] [Google Scholar]
- 35. Pappada SM, Owais MH, Cameron BD, et al. An artificial neural network-based predictive model to support optimization of inpatient glycemic control. Diabetes Technol Ther 2020; 22 (5): 383–94. [DOI] [PubMed] [Google Scholar]
- 36. Pappada SM, Cameron BD, Tulman DB, et al. Evaluation of a model for glycemic prediction in critically ill surgical patients. PLoS One 2013; 8 (7): e69475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pappada SM, Borst MJ, Cameron BD, et al. Development of a neural network model for predicting glucose levels in a surgical critical care setting. Patient Saf Surg 2010; 4 (1): 15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Van Herpe T, Espinoza M, Pluymers B, et al. An adaptive input-output modeling approach for predicting the glycemia of critically ill patients. Physiol Meas 2006; 27 (11): 1057–69. [DOI] [PubMed] [Google Scholar]
- 39. Prokhorenkova L, Gusev G, Vorobev A, Dorogush AV, Gulin A. CatBoost: unbiased boosting with categorical features. Adv Neural Inform Process Syst 2018: 6639–49. [Google Scholar]
- 40. Bojer C, Meldgaard J.. Learnings from Kaggle’s Forecasting Competitions. Work Paper 2020. [Google Scholar]
- 41. Johnson AE, Pollard TJ, Shen L, et al. MIMIC-III, a freely accessible critical care database. Sci Data 2016; 3 (1): 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Goldstein BA, Navar AM, Pencina MJ, Ioannidis J.. Opportunities and challenges in developing risk prediction models with electronic health records data: a systematic review. J Am Med Inform Assoc 2017; 24 (1): 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Ustimenko A, Prokhorenkova L, Malinin A.. Uncertainty in gradient boosting via Ensembles. arXiv Preprint 2020; Xiv–2006. ar10562. [Google Scholar]
- 44. Box GE, Cox DR.. An analysis of transformations. JRoyal Statistic///al Society: Series B (Methodological) 1964; 26 (2): 211–43. [Google Scholar]
- 45.R Core Team. R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. [Google Scholar]
- 46. Dowle M, Srinivasan A.. Data. Table: Extension of data. Frame. R Package Version 1.12 2019; 2. [Google Scholar]
- 47. Wickham H. ggplot2. Wires Comp Stat 2011; 3 (2): 180–5. [Google Scholar]
- 48. Clarke WL, Cox D, Gonder-Frederick LA, Carter W, Pohl SL.. Evaluating clinical accuracy of systems for self-monitoring of blood glucose. Diabetes Care 1987; 10 (5): 622–8. [DOI] [PubMed] [Google Scholar]
- 49. Kaufman LR, Rousseeuw PJ.. Finding Groups in Data: An Introduction to Cluster Analysis. Hoboken, NJ: John Wiley & Sons Inc;1990. [Google Scholar]
- 50. Karamched B, Hripcsak G, Albers D, Ott W.. Delay-induced uncertainty in physiological systems. arXiv Preprint ar 2020; Xiv–2007. [Google Scholar]
- 51. Ensminger SA, Morales IJ, Peters SG, et al. The hospital mortality of patients admitted to the ICU on weekends. Chest 2004; 126 (4): 1292–8. [DOI] [PubMed] [Google Scholar]
- 52. Kopecký P, Mráz M, Bláha J, et al. The use of continuous glucose monitoring combined with computer-based eMPC algorithm for tight glucose control in cardiosurgical ICU. Bio Med Res Int 2013; 2013: 1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Albers DJ, Hripcsak G, Schmidt M.. Population physiology: leveraging electronic health record data to understand human endocrine dynamics. PLoS One 2012; 7 (12): e48058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Rice MJ, Coursin DB.. Glucose meters: here today, gone tomorrow? Crit Care Med 2016; 44 (2): e97–e100. [DOI] [PubMed] [Google Scholar]
- 55. Kulkarni A, Saxena M, Price G, O’Leary MJ, Jacques T, Myburgh JA.. Analysis of blood glucose measurements using capillary and arterial blood samples in intensive care patients. Intensive Care Med 2005; 31 (1): 142–5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available at the MIMIC-III open source database at https://mimic.physionet.org/, and can be accessed following completion of required training as detailed at https://mimic.physionet.org/gettingstarted/access/.