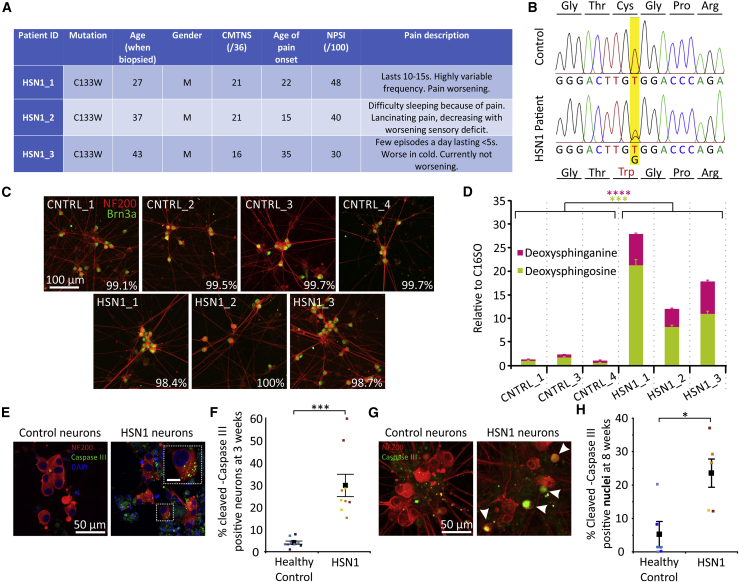
Figure 1.
HSN1 iPSC-derived sensory neurons endogenously produce neurotoxic DSBs and express high levels of caspase III
(A) Clinical phenotype of the three patients with HSN1 from which iPSCs were generated.
(B) Chromatogram showing the missense mutation 399T to G (C133W) within exon 5 of serine palmitoyl transferase, long chain base subunit-1 (SPTLC1).
(C) Representative images of iPSCdSNs from each control and HSN1 patient line used in subsequent experiments, showing NF200 (red) staining of soma and neurites and nuclear Brn3a (green) expression. Inset percentage represents cells expressing the sensory neuron marker Brn3a.
(D) Quantification of DSBs in control and HSN1 neurons matured for 4 weeks, run in triplicate. Control versus HSN1 comparison for doxSA, ∗p ≤ 0.0001 by 2-way ANOVA (F = 81.93, df = 1). Control versus HSN1 comparison for doxSO, ∗p = 0.0002 by 2-way ANOVA (F = 27.99, df = 1).
(E) Photomicrographs of 3-week-old control and HSN1 iPSCdSNs, immunocytochemically stained for NF200 and caspase III, showing the presence of caspase III “speckles” in the cytoplasm of HSN1 neurons.
(F) Quantification of neurons with speckled and nuclear caspase III expression. Control versus HSN1 comparison, ∗p = 0.0007 by 2-way ANOVA (F = 20.24, df = 1). Each data point represents the mean from an independent differentiation and iPSC line (three iPSC lines analyzed across three differentiations per genotype).
(G) Photomicrographs of 8-week-old control and HSN1 iPSCdSNs, immunocytochemically stained for NF200 and caspase III, showing the presence of caspase-III-positive nuclei in HSN1 neurons.
(H) Quantification of neurons with caspase-III-positive nuclei. ∗p = 0.016 by two-tailed Student’s t test with Welch’s correction. Each data point represents the mean from an independent differentiation and iPSC line (three iPSC lines analyzed across two differentiations per genotype, apart from HSN1_3, which was differentiated once).
Colors on graphs represent different iPSC lines, and all error bars are SEM. All images within a panel are taken at the same magnification. CMTNS, Charcot-Marie-Tooth neuropathy score (version 2); NPSI, neuropathic pain symptom inventory.