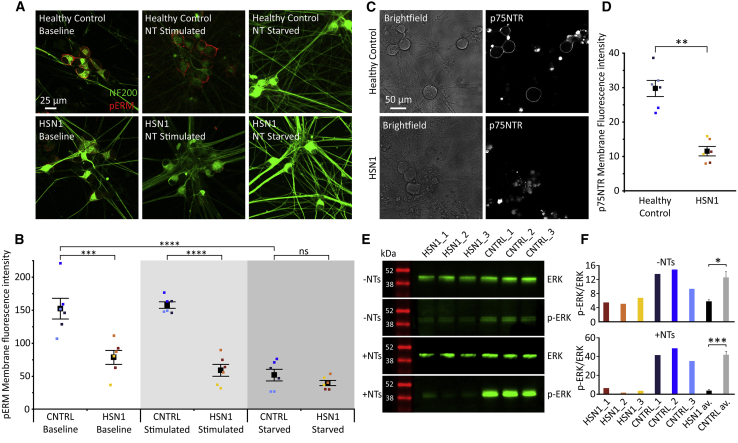
Figure 4.
Neurotrophic signaling is impaired in HSN1 neurons
(A) Photomicrographs of 8-week-old iPSCdSNs immunocytochemically stained for NF200 (green) and pERM (red) at baseline, after neurotrophic stimulation and after neurotrophic starvation.
(B) Quantification of pERM membrane fluorescence intensity determined by profile plot analysis across multiple differentiations. Between groups comparison, ∗p ≤ 0.0001 by 2-way ANOVA (F = 31.51, df = 5). Individual comparisons by Tukey’s post hoc tests: healthy control baseline versus HSN1 baseline, ∗p = 0.0001; healthy control stimulated versus HSN1 stimulated, ∗p ≤ 0.0001; healthy control starved versus HSN1 starved, p = 0.939; and healthy control baseline versus healthy control starved, ∗p = < 0.0001. Each data point represents the mean from an independent differentiation and iPSC line (three iPSC lines analyzed across two differentiations per genotype).
(C) Differential interference contrast (DIC) brightfield images of live control and HSN1 iPSCdSNs incubated with a fluorescently tagged anti-p75NTR antibody.
(D) Quantification of p75NTR membrane fluorescence intensity determined by profile plot analysis across multiple differentiations. Control versus HSN1 comparison, ∗p = 0.0003 by 2-way ANOVA (F = 38.25, df = 1). Each data point represents the mean from an independent differentiation and iPSC line (three iPSC lines analyzed across two differentiations per genotype).
(E) Western blots of 8-week-old iPSCdSNs for ERK and p-ERK with (+NT) and without (−NT) neurotrophic stimulation.
(F) Normalized relative intensity of western blots expressed as p-ERK/ERK ratio for each control and HSN1 line, and mean averages (−NT, ∗p = 0.0166 and +NT, ∗p = 0.0008 by Student’s t test).
Colors on graphs represent different iPSC lines and all error bars are SEM. All images within a panel are taken at the same magnification.