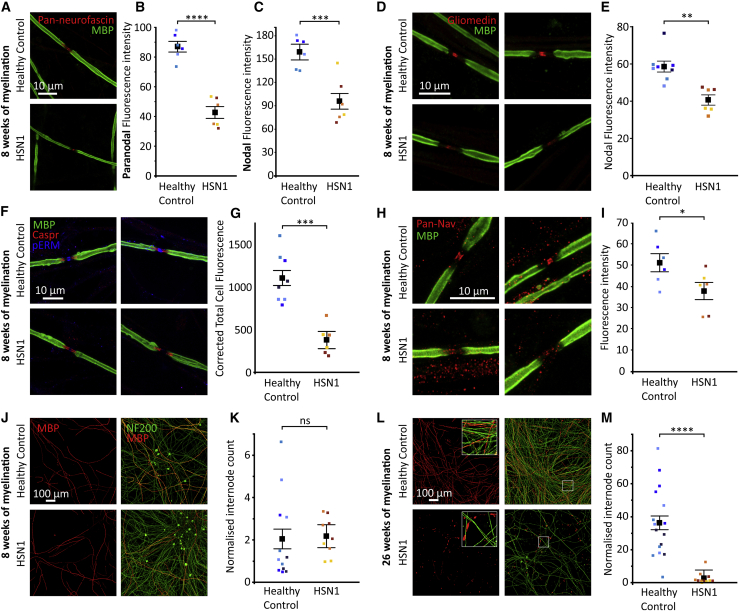
Figure 6.
Nodal and paranodal defects are observed in HSN1 myelinated neurons, with complete internode fragmentation occurring after 6 months of myelination in vitro
(A) Photomicrographs of nodes of Ranvier in myelinated cocultures immunocytochemically stained for MBP and pan-neurofascin after 8 weeks of myelination.
(B) Quantification of pan-neurofascin fluorescence intensity at the paranode determined by profile-plot analysis across multiple differentiations. Group comparison, ∗p = < 0.0001 by 2-way ANOVA (F = 74.96, df = 2) with Tukey’s post hoc tests (healthy control versus patient, ∗p = < 0.0001). Each data point represents the mean from an independent differentiation and iPSC line (three iPSC lines analyzed across two differentiations per genotype).
(C) Quantification of pan-neurofascin fluorescence intensity at the node determined by profile plot analysis across multiple differentiations. Control versus HSN1 comparison, ∗p = 0.0006 by 2-way ANOVA (F = 29.26, df = 1). Each data point represents the mean from an independent differentiation and iPSC line (three iPSC lines analyzed across two differentiations per genotype).
(D) Photomicrographs of nodes of Ranvier in myelinated cocultures immunocytochemically stained for MBP and gliomedin after 8 weeks of myelination.
(E) Quantification of nodal gliomedin fluorescence intensity determined by profile-plot analysis across multiple differentiations. Control versus HSN1 comparison, ∗p = 0.0017 by 2-way ANOVA (F = 18.02, df = 1). Each data point represents the mean from an independent differentiation and iPSC line (four control and three HSN1 iPSC lines analyzed across two differentiations per genotype).
(F) Photomicrographs of nodes of Ranvier in myelinated cocultures immunocytochemically stained for MBP, Caspr, and pERM after 8 weeks of myelination.
(G) Quantification of nodal pERM fluorescence intensity determined by profile-plot analysis across multiple differentiations. Control versus HSN1 comparison, ∗p = 0.0003 by 2-way ANOVA (F = 28.73, df = 1). Each data point represents the mean from an independent differentiation and iPSC line (four control and three HSN1 iPSC lines analyzed across two differentiations per genotype).
(H) Photomicrographs of nodes of Ranvier in myelinated cocultures immunocytochemically stained for MBP and pan-voltage gated sodium channels (pan-Nav) after 8 weeks of myelination.
(I) Quantification of nodal pan-Nav fluorescence intensity determined by profile-plot analysis across multiple differentiations. Control versus HSN1 comparison, ∗p = 0.0362 by 2-way ANOVA (F = 6.314, df = 1).
(J) Photomicrographs of myelinated co-cultures immunocytochemically stained for NF200 and MBP after 8 weeks of myelination.
(K) Quantification of internode count in control and HSN1 myelinated co-cultures after 8 weeks of myelination. Control versus HSN1 comparison (not significant) p = 0.8683 by 2-way ANOVA (F = 0.0284, df = 1). Each data point represents the mean from an independent differentiation and iPSC line (four control and three HSN1 iPSC lines were analyzed across three differentiations per genotype).
(L) Photomicrographs of myelinated cocultures immunocytochemically stained for NF200 and MBP after 26 weeks of myelination, with high magnification insets.
(M) Quantification of internode count in control and HSN1 myelinated co-cultures after 26 weeks of myelination. Control versus HSN1 comparison, ∗p = < 0.0001 by 2-way ANOVA (F = 26.47, df = 1). Each data point represents the mean from an independent differentiation and iPSC line (four control and three HSN1 iPSC lines analyzed across four differentiations per genotype, apart from HSN1_3 which was analyzed across three differentiations).
Colors on graphs represent different iPSC lines and all error bars are SEM. All images within a panel are taken at the same magnification.