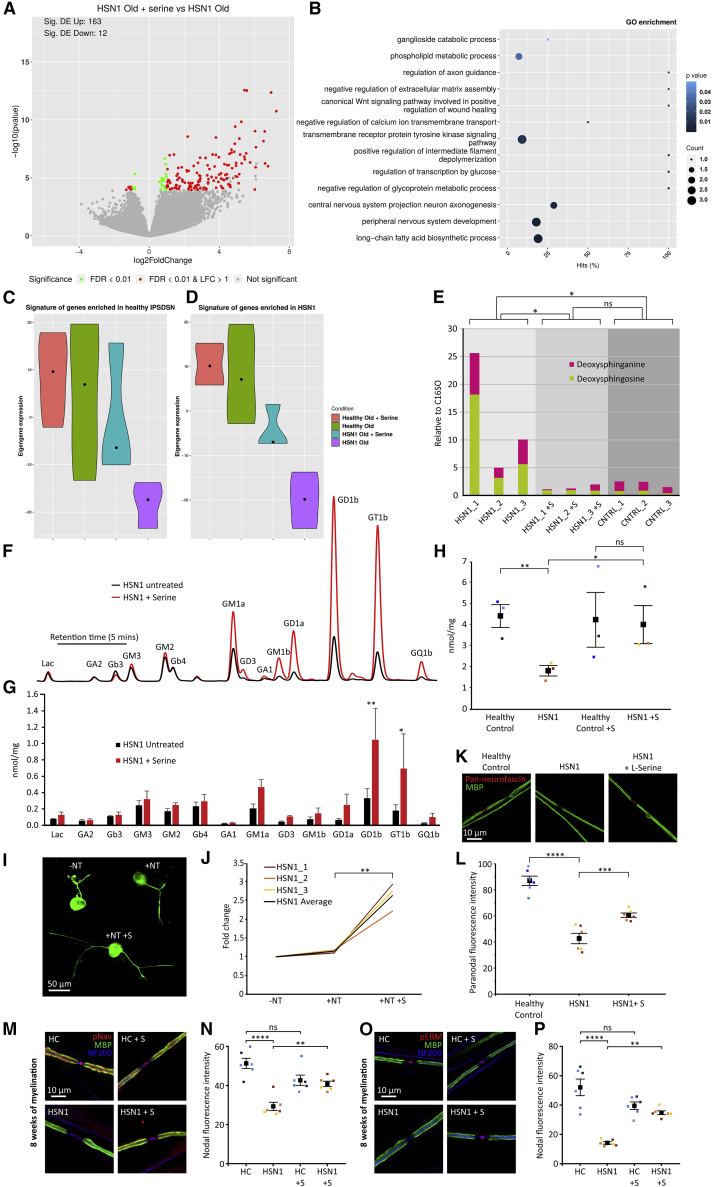
Figure 7.
l-serine treatment rescues the phenotype of HSN1 neurons and normalizes the patient transcriptome
(A) RNA-seq from 26-week-old HSN1 iPSCdSNs comparing l-serine treated to untreated. Volcano plot showing the range of differential expression of genes. Significance is color coded. The number of up/downregulated significantly DE genes is annotated in the plot.
(B) Selected significantly enriched biological processes (GO terms) in l-serine-treated HSN1 neurons versus untreated HSN1 neurons. The number of DE genes in the category is encoded in the dot size; the percentage of the category’s genes that are DE is shown on x axis, p value is color coded.
(C) Quantification of the eigengene/first principal component of genes enriched (log2 fold change < 1 and false-discovery rate [FDR] < 0.01) in control iPSCdSNs.
(D) Quantification of the eigengene/first principal component of genes enriched (log2 fold change > 1 and FDR < 0.01) in HSN1 iPSCdSNs. Violin plots show the median, interquartile range, and kernel density of the expression distribution.
(E) Quantification of deoxysphingoid bases (DSBs) in control, HSN1, and HSN1 + l-serine (HSN1 + S)-treated neurons matured for 8 weeks. DoxSA/DoxSO combined group comparison, ∗p = 0.0021 by 2-way ANOVA (F = 8.842, df = 2) with Tukey’s post hoc test for individual comparisons. ∗p = 0.018 for control versus HSN1; ∗p = 0.012 for HSN1 versus HSN1 + S; p = 0.981 for control versus HSN1 + S.
(F) HPLC profile plots of glycosphingolipids from representative HSN1 untreated and HSN1 with l-serine treatment neurons at 8 weeks after differentiation.
(G) Quantification of individual glycosphingolipids normalized to total protein input. Group comparison, ∗p = 0.0011 by 2-way ANOVA (F = 5.747, df = 3) with Tukey’s post hoc for individual GSL comparison. Each bar represents the mean from three separate control or HSN1 lines.
(H) Total glycosphingolipid levels normalized to protein input. Group comparison, p = 0.0011 by 2-way ANOVA (F = 5.747, df = 3) with Tukey’s post hoc tests (HSN1 versus HSN1 + S, ∗p = 0.0151).
(I) Photomicrographs of 26-week-old HSN1 iPSCdSNs 24 h after dissociation and replating, immunocytochemically stained for NF200, showing treatment groups: without neurotrophic factors (−NT), with neurotrophic factors (+NT), and with neurotrophic factors and l-serine (+NT +S).
(J) Quantification of total neurite length showing fold change between treatment groups. ∗p = 0.0024 by 2-tailed unpaired t test.
(K) Photomicrographs of nodes of Ranvier after 8 weeks of myelination, immunocytochemically stained for MBP (green) and pan-neurofascin (red), showing myelinated cultures from healthy control, HSN1, and HSN1 with l-serine groups.
(L) Quantification of pan-neurofascin paranodal fluorescence intensity determined by profile-plot analysis across multiple differentiations. Group comparison, ∗p = < 0.0001 by 2-way ANOVA (F = 74.96, df = 2) with Tukey’s post hoc tests (healthy control versus patient, ∗p = < 0.0001; HSN1 versus HSN1 + S, ∗p = 0.001). Each data point represents the mean from an independent differentiation and iPSC line (three iPSC lines analyzed across two differentiations per genotype). Colors on graphs and profile plots represent different iPSC lines.
(M) Photomicrographs of nodes of Ranvier after 8 weeks of myelination, immunocytochemically stained for MBP (green), pan-Nav (red), and NF200 (blue), showing myelinated cultures from healthy control, healthy control with l-Serine, HSN1, and HSN1 with l-serine.
(N) Quantification of pan-Nav fluorescence intensity determined by profile-plot analysis across two differentiations. Group comparison, ∗p = < 0.0001 by 2-way ANOVA (F = 17.37, df = 3) with Tukey’s post hoc tests (healthy control versus patient, ∗p = < 0.0001; HSN1 versus HSN1 + S, ∗p = 0.0086). Each data point represents the mean from an independent differentiation and iPSC line (three iPSC lines analyzed across two differentiations per genotype).
(O) Photomicrographs of nodes of Ranvier after 8 weeks of myelination, immunocytochemically stained for MBP (green), pERM (red), and NF200 (blue), showing myelinated cultures from healthy control, healthy control with l-serine, HSN1, and HSN1 with l-serine.
(P) Quantification of pERM fluorescence intensity determined by profile plot analysis across two differentiations. Group comparison, ∗p = < 0.0001 by 2-way ANOVA (F = 21.08, df = 3) with Tukey’s post hoc tests (healthy control versus patient, ∗p = < 0.0001; HSN1 versus HSN1 + S, ∗p = 0.0038). Each data point represents the mean from an independent differentiation and iPSC line (three iPSC lines analyzed across two differentiations per genotype).
Colors on graphs and profile plots represent different iPSC lines and all error bars are SEM. All images within a panel are taken at the same magnification.