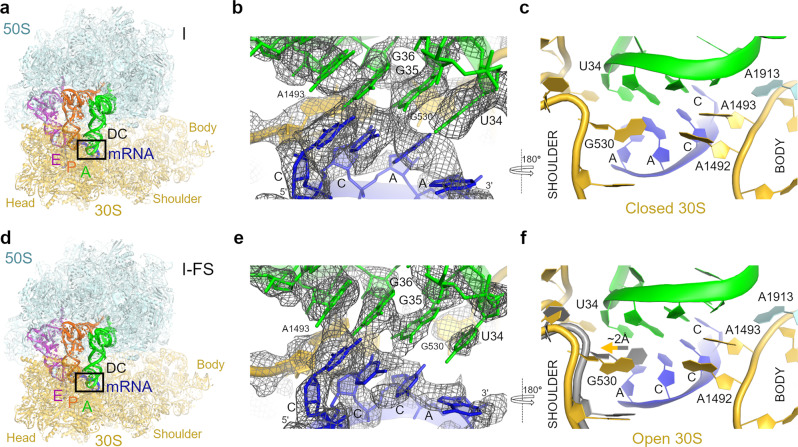
Fig. 3. Cryo-EM structures of pre-translocation 70S formed with fMet-tRNAfMet (P site) and Pro-tRNAPro (A site).
a Overall view of the 70S structure with non-frameshifting mRNA (CCA-A; Structure I). Weaker density in the E-site than in the A and P sites suggests partial occupancy of E-tRNA (“Methods”). b Cryo-EM density (gray mesh) for codon–anticodon interaction between non-frameshifting mRNA and tRNAPro in the A site of Structure I. The view approximately corresponds to the boxed decoding center region (DC) in (a). The map was sharpened with a B-factor of -80 Å2 and is shown at 2.5 σ. c Decoding center nucleotides G530 (in the shoulder region) and A1492-A1493 (in the body region) stabilize the codon–anticodon helix in Structure I. d Overall view of the 70S structure with the slippery mRNA (CCC-A; Structure I-FS). Weaker density in the E-site than in the A and P sites suggests partial occupancy of E-tRNA (see “Methods”). e Cryo-EM density (gray mesh) for codon–anticodon interaction between the slippery mRNA codon and tRNAPro in Structure I-FS. The map was sharpened with a B-factor of -80 Å2 and is shown at 2.5 σ. f Partially open conformation of the 30S subunit due to the shifted G530 (in the shoulder region) in Structure I-FS relative to that in Structure I (16S shown in gray). Structural alignment was obtained by superposition of 16S ribosomal RNAs (rRNAs). In all panels, the 50S subunit is shown in cyan, 30S subunit in yellow, mRNA in blue, tRNAPro in green, tRNAfMet in orange and sub-stoichiometric E-site tRNA in magenta.