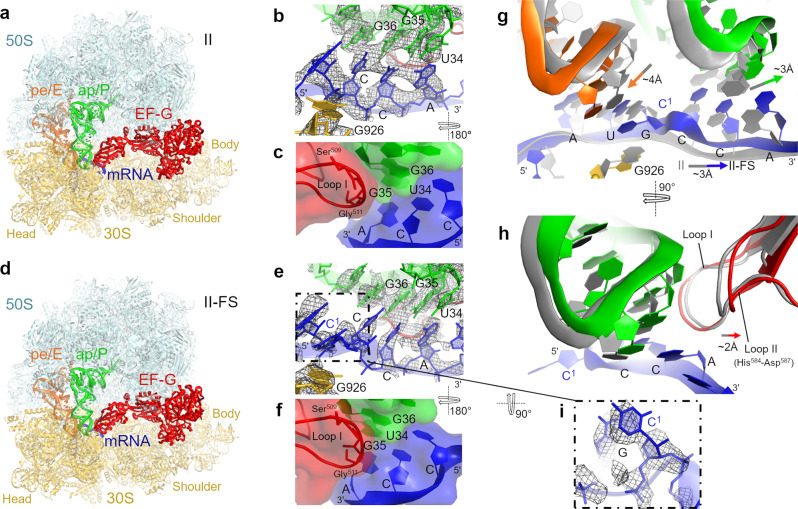
Fig. 4. Cryo-EM structures of mid-translocation states formed with EF-G•GDPCP.
a Overall view of mid-translocation Structure II with the non-frameshifting mRNA. b Cryo-EM density (gray mesh) of the non-frameshifting tRNAPro and mRNA codon near the P site. The map is sharpened by applying the B-factor of -80 Å2 and is shown with 2.5 σ. c Interaction of the EF-G loop I (Ser509-Gly511, red) with the codon–anticodon helix (space-filling surface and cartoon representation). d Overall view of mid-translocation Structure II-FS with the frameshifting mRNA. e Cryo-EM density (gray mesh) of the frameshifting tRNAPro and mRNA codon near the P site. The map is sharpened by applying the B-factor of -80 Å2 and is shown with 2.5 σ. Note the unpaired and bulged C1 nucleotide in the mRNA, also shown in (i). f Interaction of the EF-G domain IV loop I (Ser509-Gly511) with the codon–anticodon helix of the frameshifting mRNA (compare to c). g Differences in positions of tRNAPro (green) and tRNAfMet (orange) in the frameshifting structure II-FS relative to those in the non-frameshifted structure II (gray). h Adjustment of loop II of domain IV of EF-G (red) to accommodate the shifted position of tRNAPro (green) in the frameshifting structure II-FS relative to those in structure II (gray). Structural alignments were performed by superposition of 16S rRNAs. i Close-up view of cryo-EM density for bulged C1 in Structure II-FS (also shown in e). The ribosomal subunits, tRNAs, and mRNA are colored as in Fig. 3, EF-G is shown in red.