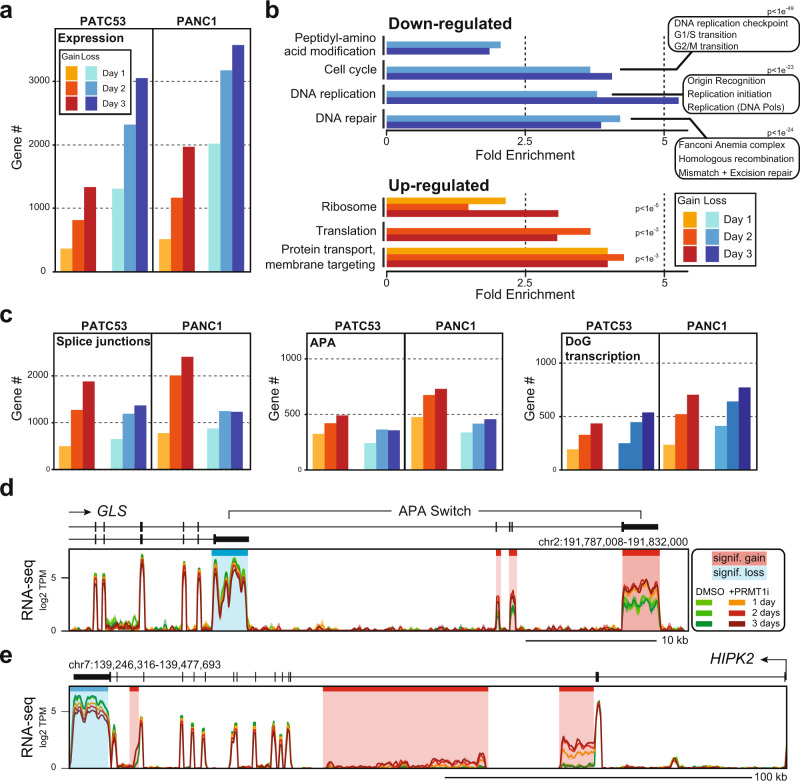
Fig. 4. PRMT1 plays a critical role in regulating gene expression and co-transcriptional RNA processing.
a Number of genes showing significant gain or loss of gene expression in PATC53 and PANC1 cells treated with 1 µM PRMTi for 1, 2, and 3 days. b Significantly enriched gene ontology (GO) categories (PANTHER) for down or upregulated genes as a function of time after PRMTi treatment in the cells described in (a). Key GO categories and p-values of enrichment are indicated. c Number of genes showing significant gain or loss of splicing junctions, alternative polyadenylation (APA), and downstream of gene (DoG) transcription events in the cells described in (a). d Representative composite screenshots of RNA-seq data over GLS, a gene that undergoes a treatment- and time-specific APA switch between two annotated 3’-UTRs, in PANC1 cells. e Representative composite screenshots of RNA-seq data over HIPK2, a gene that similarly to GLS undergoes significant APA gains over previously unannotated 3’-UTRs (shaded red) and concomitant loss of distal 3’-UTR usage (shaded blue), in PATC53 cells. Data from control and PRMTi-treated cells are color-coded as indicated.