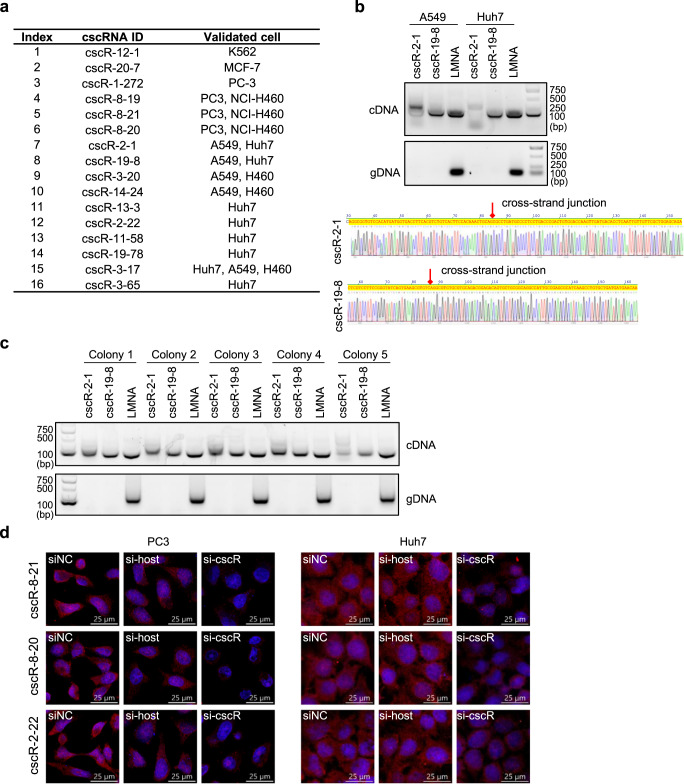
Fig. 3. Experimental validations of the cscRNAs.
a 16 cscRNAs were experimentally validated in different cell lines, by RT-PCR followed by Sanger sequencing. b As two examples, the sequences covering the cross-strand junction sites of cscR-2-1 and cscR-19-8 were amplified by PCR with the cDNA of A549 and Huh7 cells (top). The amplified sequences were subjected to Sanger sequencing (bottom). The Sander sequencing results of the other 14 cscRNAs are shown in Fig. S8. Source data are provided as a Source data file. c The sequences covering the cross-strand junction sites of cscR-2-1 and cscR-19-8 were amplified by PCR with the cDNA (top) or genomic DNA (bottom) from 5 cell colonies derived from A549 single cells. Representative images from three repeated experiments are shown. Source data are provided as a Source data file. d Representative images of RNA FISH, from three biological replicates, with the probes targeting cscR-8-21, cscR-8-20, or cscR-2-22 (red) in PC3 and Huh7 cells. The cell nucleus was stained with DAPI (blue). RNA knockdown with the siRNAs targeting the cscRNAs or the potential host gene transcripts were performed to show the specificity of the FISH probes.