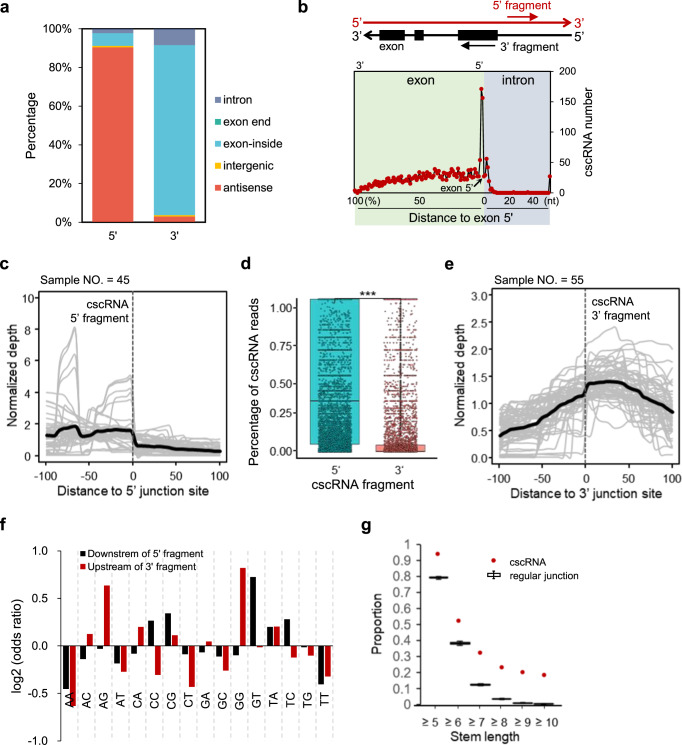
Fig. 6. Features of the cross-strand junctions related to cscRNA biogenesis.
a Proportions of the cscRNAs categorized according to the genomic annotations of the 5′ or 3′ junction sites. antisense: the antisense strand of an annotated gene; intergenic: intergenic region of the genome not being annotated to any gene; exon-inside: inside of an exon region; exon end: the 5′ cross-strand junction site being the 3′ end of an exon or the 3′ junction end being the 5′ end of an exon; intron: inside of an annotated intron. b Distributions of the distances between the 3′ junction sites and the closest exon 5′ ends. Note that for the 3′ junction sites located inside of an exon (left part of the plot), their distances to the exon 5′ ends were transformed into percentages via normalization by the length of the hosting exons. For the junction sites located in the introns, the distances (nt) to the closest downstream exon 5′ ends were recorded (right part). c Average read depths along the genome around the 5′ junction sites (200 nt in total). For each sample (represented by a gray line), the value of each position was calculated by taking the average of the read depths of the multiple cscRNAs expressed in the particular sample. All the values along the 200-nt region were then normalized by their average, so that the different lines, indicating different samples, are in the same normalized scale. To control the stochastic noise, we only used the 5′ junction sites around which the average read depths within the 200-nt region were not lower than 5. Such analyses were performed for 45 samples, which have at least 5 cscRNAs with the 5′ junction sites passing the filter above. Finally, the average of the 45 samples on each position was shown as the thick black line. d The RNA-seq reads covering each of the 5′ or 3′ cross-strand junction sites in each sample are composed of two types of the reads, i.e., the reads that supported the cscRNAs and the reads that were mapped to the genome and therefore supporting the regular RNA transcripts. The proportions of the RNA-seq reads that supported the cscRNAs were summarized as box plots for the 5′ and 3′ junction sites of all the recurrent cscRNAs (Two-sided t-test, p-value = 2.2e−16). The median value is shown as the line and the average as the cross. e Average read depths along the genome around the 3′ junction sites (200 nt in total). The lines were prepared with the same method in panel d, for 55 samples, which have at least 5 cscRNAs with the 3′ junction sites passing the filter. Finally, the average of the 55 samples on each position was shown as the thick black line. f Odds ratios of the dinucleotide motifs at the downstream of the 5′ junction sites and the upstream of the 3′ junction sites of the recurrent cscRNAs. 10,000 randomly picked positions of the genome was used as the background. g The percentages of the recurrent cscRNAs that have complementary sequences between their 5′ and 3′ fragments. As a reference, the probabilities of detecting complementary sequences between the up- and downstream regions of 10,000 randomly selected exon–exon junction sites were counted. Such randomization was performed for 100 times, and the 100 probabilities were summarized as box plots (Two-sided t-test P-value from left to right: 1.08E−73, 1.11E−61, 7.76E−92, 2.02E−115, 6.92E−120, 6.17E−130). The median value is shown as the line and the average as the cross. Source data are provided as a Source data file.