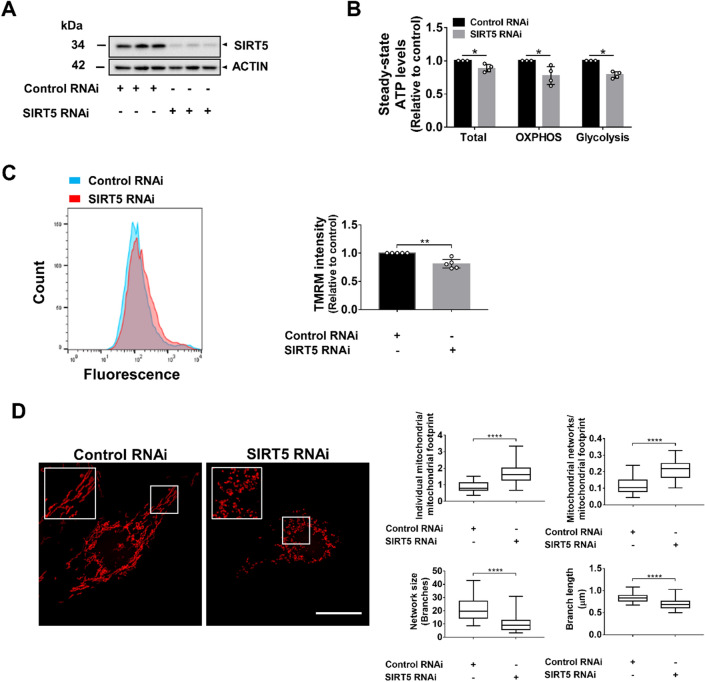
Figure 2.
SIRT5-knockdown impairs cellular energy metabolism and induces mitochondrial fragmentation. SIRT5-knockdown by RNAi was carried out in hPTECs. Non-targeting siRNA served as control. All experiments were performed 72 h after siRNA transfection. (A) WB showing SIRT5 protein levels. Whole WB shown in supplementary file (Supplementary Fig. S8b). (B) Bar graph shows ATP levels. hPTECs (control and SIRT5 RNAi) were analyzed without inhibitor treatment to determine total ATP levels (glycolytic + mitochondrial ATP), after treatment with 2-deoxyglucose to inhibit glycolysis (mitochondrial ATP; OXPHOS) or after treatment with oligomycin A to inhibit mitochondrial ATP synthase (glycolytic ATP; Glycolysis). ATP levels were normalized to cell number as measured by DNA quantification. Data are from four independent experiments with n = 5–6 technical replicates/group. (C) Bar graph showing TMRM fluorescent intensity. hPTECs (control and SIRT5 RNAi) were incubated with TMRM (25 nM), a fluorescent dye which accumulates in mitochondria based on the ΔΨM and fluorescent intensities were determined by flow cytometry. Background fluorescence was assessed by FCCP treatment (10 µM) prior to TMRM incubation and fluorescence intensities were subtracted from the TMRM fluorescence intensities. Data are from five independent experiments with n = 2 technical replicates/group. Data are median ± IQR. To determine statistical significance a Mann–Whitney U test was applied. *p < 0.05 and **p < 0.01. (D) Cells were stained with Mitotracker Red CMXRos (100 nM) and imaged by confocal microscopy. Scale bar: 20 µm. (B) Mitochondrial morphology was quantitatively assessed using the Mitochondrial Network Analysis (MiNA) toolset (ImageJ). The number of (i) individual mitochondria and (ii) mitochondrial networks per cell were quantified and normalized to the mitochondrial footprint; (iii) mean network size (number of branches) and the mean branch length (µm) were determined. Forty-two control RNAi-treated and 51 SIRT5 RNAi-treated cells from three independent experiments were analyzed. Box and whisker plots display median interquartile range and minimum and maximum values. Mann–Whitney U test was used to determine statistical significance. ****p < 0.0001.