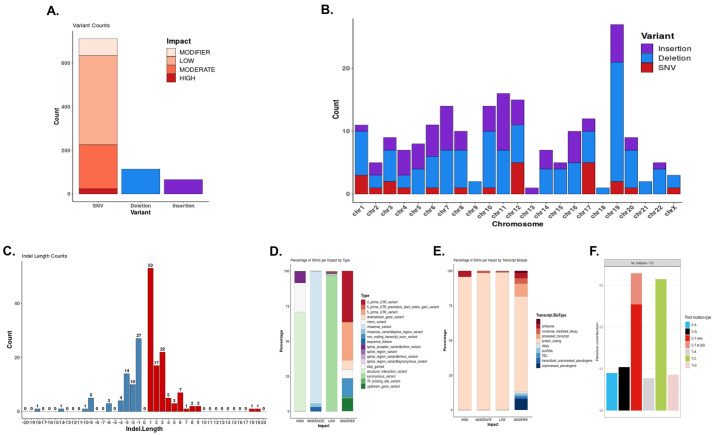
Fig. 5.
Variants in EMM PDOX detected by whole-exome sequencing. (A) Total number of SNVs classified as high, moderate and low impact and modifier, and insertions and deletions. (B) Variant counts per chromosome in terms of high-impact SNVs, insertions and deletions (indels). (C) Counts of indels by length. In the analysis, indels were considered as deletions or insertions of up to 20 bp. (D,E) Percentage of SNVs per impact by type (D) and by transcript biotype (E). All 712 SNVs were included in the analysis. (F) Relative concentrations of point mutation types detected. To obtain the COSMIC signatures contribution, it was assumed that all SNVs detected corresponded to somatic mutations.