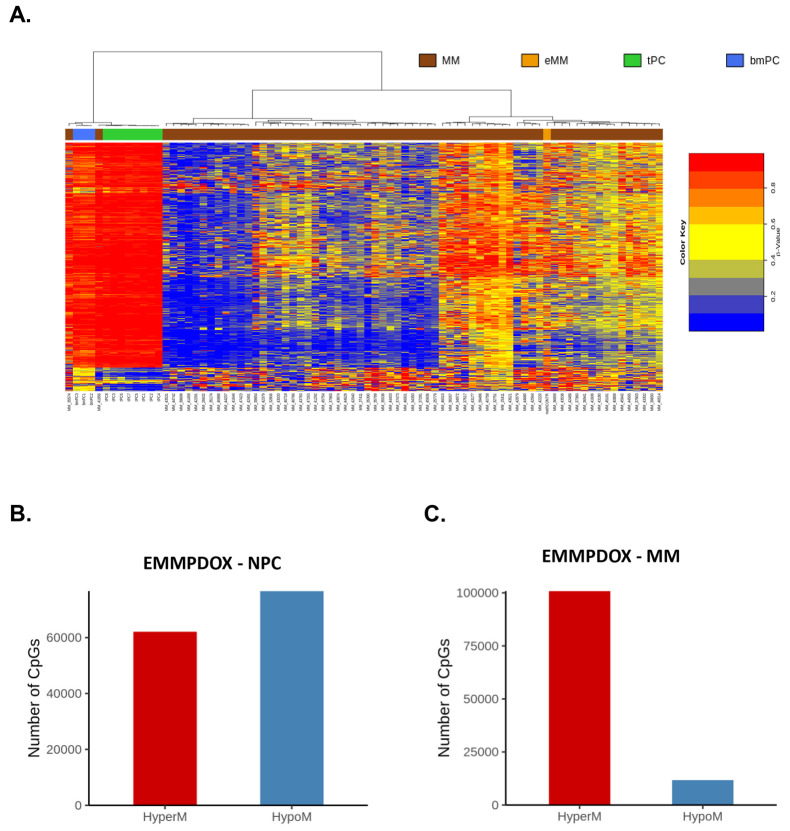
Fig. 6.
Whole-genome methylation analysis of EMM PDOX. (A) Unsupervised hierarchical clustering analysis of EMM PDOX, MM, NPCs from tonsil (tPC) or NPCs from bone marrow (bmPC). The 5000 CpG sites with the most variable methylation values were used. (B) Differentially methylated CpGs in EMM PDOX compared to NPCs and (C) newly diagnosed and chemotherapy-naïve MM patients (Agirre et al., 2015). Compared with NPCs, we identified 138,645 differentially methylated CpGs, including 55.3% hypomethylated and 44.7% hypermethylated. As compared with MM, we identified 112,551 differentially methylated CpGs, including 10.4% hypomethylated and 89.6% hypermethylated. Epigenetic data were obtained using an Infinium MethylationEPIC BeadChip array (Illumina) for EMM PDOX (from four different fragments of the tumor) and using a 450 K methylation array (Illumina) for MM and NPCs (Agirre et al., 2015). To compare the results with the published 450 K methylation arrays, only the common probes with EPIC were taken into account, and average methylation values were used for each group.