Figure 4 .
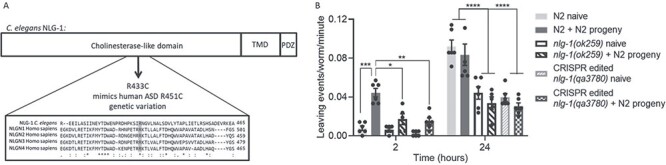
R433C mutation in nlg-1 phenocopies social impairment of nlg-1 null. (A) Domain structure of NLG-1 indicating the arginine to cysteine (R-C) amino acid substitution at position 433 generated within the cholinesterase like domain following CRISPR/Cas9. Protein sequence alignment of C. elegans NLG-1 and human NLGN1–4 indicates the arginine residue involved in ASD and its conservation in C. elegans (shaded in grey). ‘*‘Indicates conservation of a single amino acid residue, ‘:’ indicates conservation between amino acid groups with similar properties and ‘.’ indicates conservation between amino acid groups with weakly similar properties. (B) The number of food-leaving events were counted for N2, nlg-1(ok259) and CRISPR generated nlg-1(qa3780) on naïve and N2 progeny conditioned food lawns. Data are mean ± SEM. N2, nlg1(ok259) and nlg-1(qa3780) n = 6. Two-way ANOVA with Tukey’s multiple comparison test; *P < 0.05, **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001.
