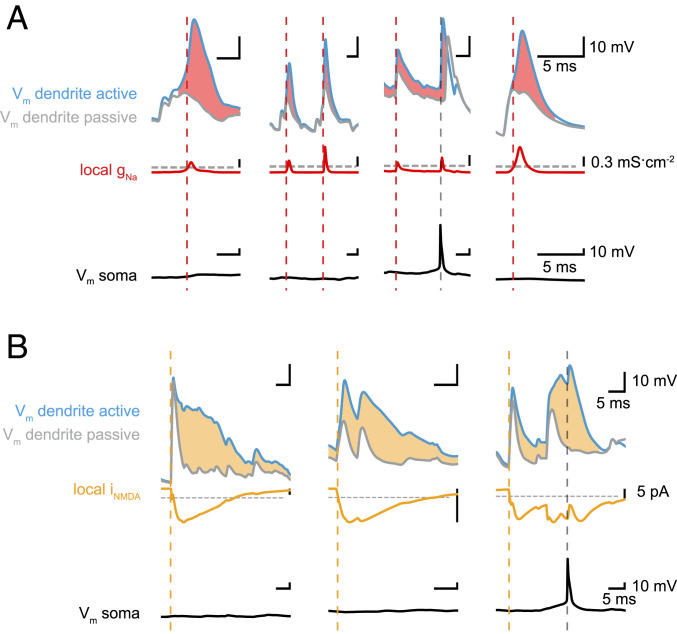
Fig. 2.
Detecting dendritic Na+ spikes and NMDA spikes. (A) Four examples of local dendritic Na+ spikes showing dendritic membrane potential (Top), dendritic Na+ conductance (Center), and somatic membrane potential (Bottom). We compared simulations in a fully active (light blue) and a passive dendrite (gray) with identical synaptic input. Dendritic Na+ spikes (initiation indicated by dashed red lines) are characterized by a >10% voltage amplitude difference (light red shading) between active and passive simulations. We use a threshold (gray horizontal dashed line) on the local Na+ conductance (Center, red) as a proxy to efficiently detect dendritic Na+ spikes in all following simulations. By simultaneously monitoring somatic Vm, we can distinguish local events from bAPs (gray vertical dashed lines; Materials and Methods). (B) Three examples of local dendritic NMDA spikes showing dendritic membrane potential (Top), local NMDA current (Center), and somatic membrane potential (Bottom). We compared simulations in a dendrite with voltage-dependent NMDA conductances (light blue) and the same dendrite with NMDA conductances frozen at the resting potential (gray), with identical synaptic input. Dendritic NMDA spikes (initiation indicated by dashed orange lines) are characterized by a >10% voltage amplitude difference (light orange shading) between active and passive simulations. We use a threshold (gray horizontal dashed line) on the local NMDA current (Center, orange) and a threshold on the local dendritic membrane potential, as well as a threshold on the total charge delivered during a spike, as a proxy to efficiently detect dendritic NMDA spikes in all following simulations (Materials and Methods).