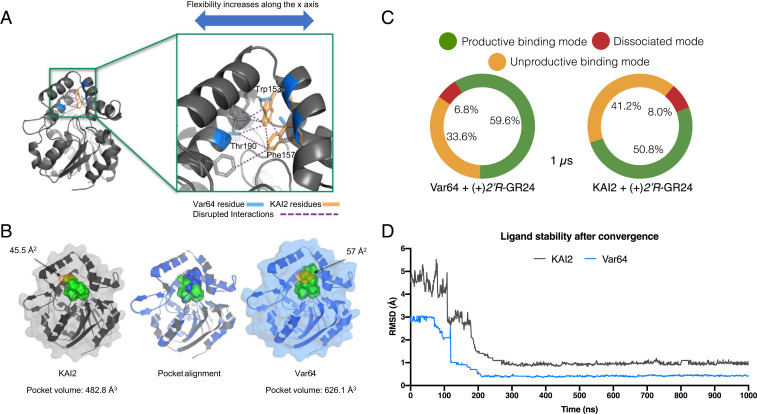
Fig. 4.
Var64-derived amino acids create an SL-binding domain. (A) The substitution of KAI2 aromatic residues for ShHTL7 equivalents results in the disruption of several interactions (purple dashed lines) along the x-axis of the protein. This disruption translates into increased motility of the lid domain (Movie S1), consistent with the receptor’s new ability to accommodate SL molecules. (B) Pocket alignment of KAI2 and Var64 receptors reveals an 11.5 Å larger pocket mouth (orange shading) and a 29.7% increased pocket volume in the Var64 receptor. The pocket mouths are shown in orange shading, with width shown in Å2. (C) A summary of the three most common binding states between Var64, KAI2, and 2′R-GR24. Binding modes of ligand–protein complexes were determined by 1-µs MD simulations of Var64 and KAI2 receptors. The Var64–(2′R)-GR24 complex was found in a productive binding mode 59.6% of the time, while the same state was found only 50% of the time for the KAI2 complex. (D) Calculation of the magnitude change in the pose of the ligand throughout the MD simulation. Both proteins stabilized the ligand after 0.1-µs simulation, but its position on the KAI2 pocket is compromised briefly before final convergence, as the interaction is less stable.