Fig. 4.
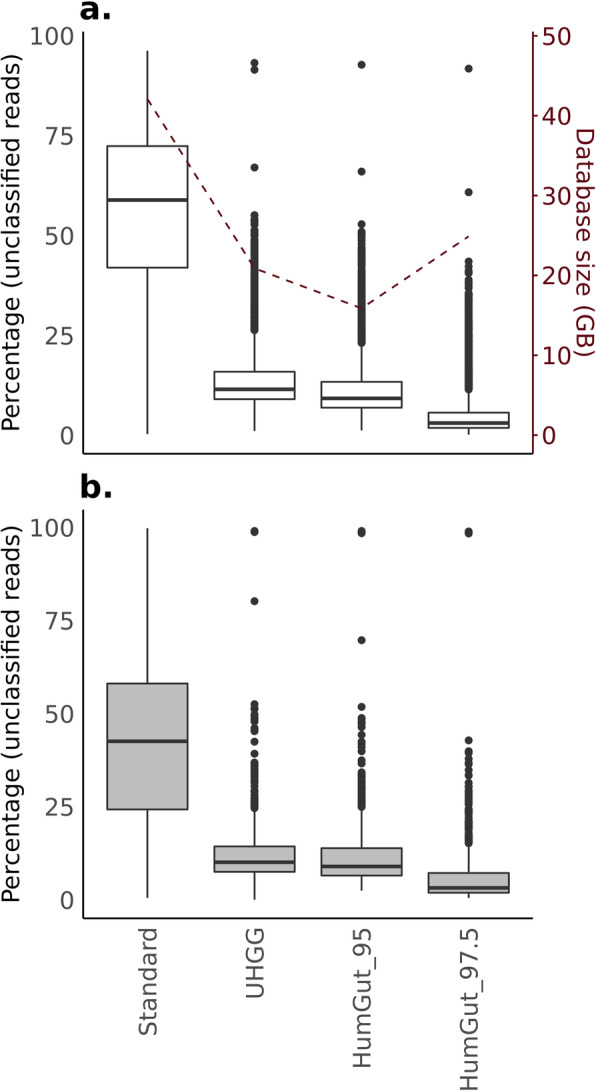
The performance of HumGut versions in comparison to the standard Kraken2 database and UHGG collection. a. The boxplot shows the distribution of unclassified reads for the 3,534 analyzed healthy reference metagenome samples. The dashed line represents the k-mer database sizes (right y-axis). Every database version includes standard human genome sequences, in addition to database-specific (sub)sets of bacteria and archaea, and the difference in size is only due to differences in the latter. b The classification of an additional 963 human gut metagenomes, not part of the reference set
