Significance
SNF1-related protein kinase 2s (SnRK2s) are central regulators in abscisic acid (ABA) signaling and stress responses in plants. However, SnRK2 substrates are largely unknown. Here, we identified Arabidopsis group C Raf-like kinases Raf36 and Raf22 as SnRK2 substrates. Reverse genetic analyses showed that Raf36 and Raf22 negatively regulate ABA signaling. Under optimal growth conditions, Raf36 accumulates in the cell and phosphorylates substrates to suppress ABA responses. Conversely, under abiotic stress conditions, ABA-activated SnRK2s phosphorylate Raf36 and promote Raf36 degradation, leading to stronger ABA responses. Taken together, this study provides insight into molecular and physiological functions of group C Raf kinases in ABA signaling.
Keywords: abscisic acid, Raf-like kinase, SnRK2, Arabidopsis thaliana
Abstract
The phytohormone abscisic acid (ABA) plays a major role in abiotic stress responses in plants, and subclass III SNF1-related protein kinase 2 (SnRK2) kinases mediate ABA signaling. In this study, we identified Raf36, a group C Raf-like protein kinase in Arabidopsis, as a protein that interacts with multiple SnRK2s. A series of reverse genetic and biochemical analyses revealed that 1) Raf36 negatively regulates ABA responses during postgermination growth, 2) the N terminus of Raf36 is directly phosphorylated by SnRK2s, and 3) Raf36 degradation is enhanced in response to ABA. In addition, Raf22, another C-type Raf-like kinase, functions partially redundantly with Raf36 to regulate ABA responses. A comparative phosphoproteomic analysis of ABA-induced responses of wild-type and raf22raf36-1 plants identified proteins that are phosphorylated downstream of Raf36 and Raf22 in planta. Together, these results support a model in which Raf36/Raf22 function mainly under optimal conditions to suppress ABA responses, whereas in response to ABA, the SnRK2 module promotes Raf36 degradation as a means of alleviating Raf36-dependent inhibition and allowing for heightened ABA signaling to occur.
Environmental stresses, such as drought, high salinity, and low temperature, have adverse effects on plant growth and development. Abscisic acid (ABA) is a phytohormone that plays important roles in responses and adaptations to these stresses, as well as in embryo maturation and seed dormancy (1, 2). The major ABA signaling pathway consists of three core components: ABA receptors, type 2C protein phosphatases (PP2Cs), and SNF1-related protein kinase 2s (SnRK2s) (3, 4). In this pathway, SnRK2s transmit ABA- or osmostress-induced signals through phosphorylation of downstream substrates, thereby promoting ABA- or stress-inducible gene expression and stomatal closure (5–8). The Arabidopsis genome contains 10 members of SnRK2, and they are classified into three subclasses (9, 10). Among them, subclass III members, SRK2D/SnRK2.2, SRK2E/OST1/SnRK2.6, and SRK2I/SnRK2.3, are essential for ABA responses (11–14).
Raf-like protein kinases were recently identified as regulators of ABA signaling. Among 80 putative mitogen-activated protein kinase kinase kinases (MAPKKKs) in Arabidopsis, 48 members are categorized as Raf-like subfamilies and can further be divided into 11 subgroups (B1 to B4 and C1 to C7) (15). In Physcomitrella patens, the ABA and abiotic stress-responsive Raf-like kinase (ARK) gene, also named as ABA nonresponsive (ANR) or constitutive triple response 1-like (CTR1L), is required for ABA-responsive SnRK2-activation, gene expression and drought, osmotic and freezing tolerance (16–18). ARK encodes a B3 subgroup Raf-like protein kinase that phosphorylates SnRK2s in vitro, suggesting that ARK functions as an upstream kinase of SnRK2s (16). In addition, several recent studies also confirmed that Arabidopsis B2, B3, and B4 subgroup of Raf-like kinases are required for osmotic stress–induced SnRK2 activation, gene expression, and stomatal closure (19–23). In addition to group B, it had been reported that several group C Raf-like kinases are associated with ABA responses. For example, Arabidopsis Raf43, a C5 kinase, regulates ABA sensitivity during seed germination and seedling root growth (24), whereas Raf22, a member of C6 subgroup, negatively regulates stress- or ABA-induced growth arrest (25). However, despite these ABA-related genetic phenotypes, it is still unclear whether group C kinases directly regulate SnRK2-dependent signaling pathways.
In this study, we identified Raf36, a C5 group Raf-like kinase, as a protein that directly interacts with, and is phosphorylated by, SnRK2. Our evidence indicates that Raf36 functions as a negative regulator of ABA signaling pathway during the postgerminative growth stage under the control of SnRK2. In addition, we revealed that Raf22, a C6 Raf-like kinase, functions partially redundantly with Raf36. Raf36 mainly functions under optimal conditions, because its degradation was promoted under ABA treatment. Furthermore, comparative phosphoproteomic analysis unveiled the phosphorylation network regulated by Raf22 and/or Raf36 in planta. Collectively, unlike group B Rafs, which have been recently reported as an “accelerator” of ABA response upstream of SnRK2s, our results demonstrate that Arabidopsis group C Rafs, Raf22 and Raf36, function as a “brake” of ABA response downstream of SnRK2s.
Results
Raf36 Interacts with Subclass III SnRK2.
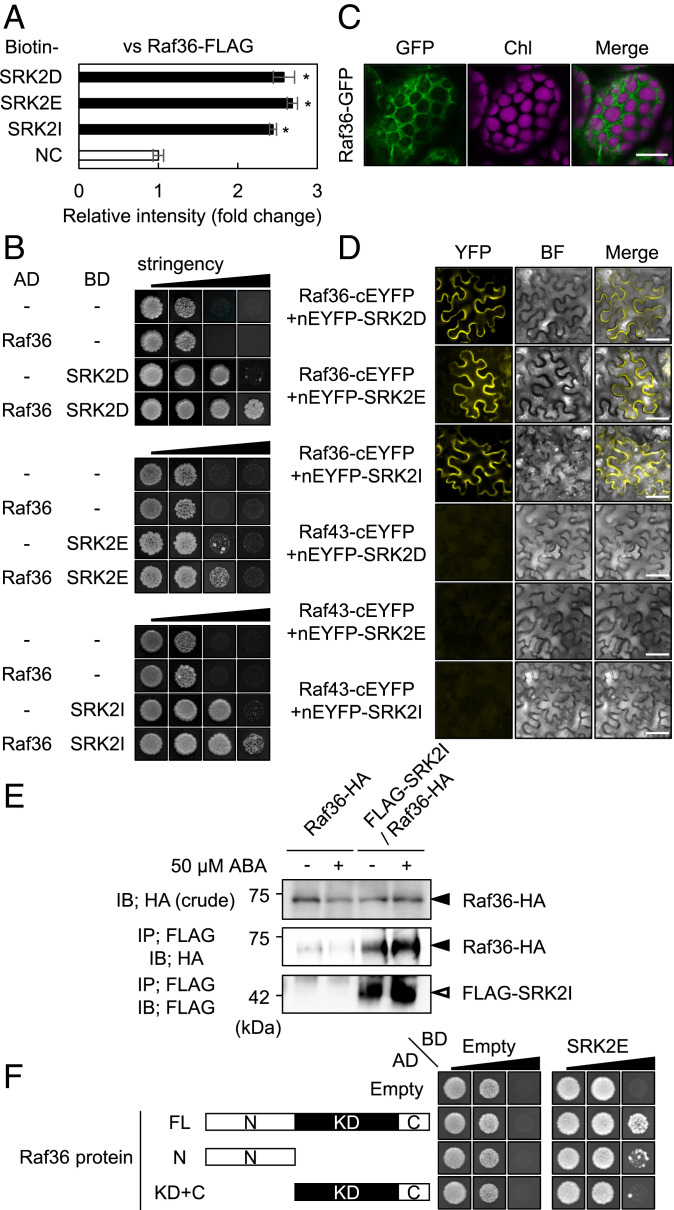
To identify additional kinases that regulate ABA signaling pathways, we used the AlphaScreen assay to screen a collection of Arabidopsis MAPKKK proteins for their ability to physically interact with SRK2I (SnRK2.3), a subclass III SnRK2. From a pilot experiment, several Raf-like protein kinases were identified as candidate interactors with SRK2I (SI Appendix, Fig. S1 A–C). Raf36, which belongs to a C5 subgroup kinase (SI Appendix, Fig. S2), was one of the SRK2I-interacting proteins. Interaction between Raf36 and SRK2I, as well as between Raf36 and additional subclass III SnRK2s, SRK2D (SnRK2.2), and SRK2E (OST1/SnRK2.6), was confirmed by AlphaScreen assay (Fig. 1A) and yeast two-hybrid assay (Fig. 1B). SnRK2s were previously found within the cytosol and nuclei of Arabidopsis cells (14), and we observed that Raf36-GFP is localized mainly in the cytosol (Fig. 1C). To determine in vivo interaction between Raf36 and SnRK2s, bimolecular fluorescence complementation (BiFC) assay was performed. A strong YFP fluorescence signal was observed in the cytosol when Raf36 was coexpressed with SRK2D, SRK2E, or SRK2I, confirming their interaction in planta (Fig. 1D and SI Appendix, Fig. S3). On the other hand, no signal was observed when Raf43, a homolog of Raf36, was employed as a negative control protein (Fig. 1D and SI Appendix, Fig. S3). Furthermore, coimmunoprecipitation assays demonstrated that Raf36 interacted with SRK2I in Arabidopsis cells under both control and ABA-treated conditions (Fig. 1E). Together, these results demonstrate that Raf36 physically interacts with ABA-responsive SnRK2s both in vitro and in vivo.
Fig. 1.
Raf36 interacts with subclass III SnRK2s. (A) AlphaScreen assay shows interaction of Raf36 and subclass III SnRK2s. Bars indicate means ± SE (n = 3), and asterisks indicate significant differences by two-tailed Student’s t test (P < 0.05). (B) Yeast two-hybrid (Y2H) assay shows interaction between Raf36 and subclass III SnRK2s. Yeast cells expressing GAL4AD:Raf36 and GAL4BD:SnRK2s fusion proteins were incubated on SD media supplemented with or without 3-amino-1,2,4-triazole (3-AT) and lacking combinations of amino acids leucine (L), tryptophan (W), and histidine (H), as follows (in order from low to high stringency): -LW, -LWH, -LWH +10 mM 3-AT, -LWH +50 mM 3-AT. Photographs were taken at 10 d (SRK2D and SRK2E) or 12 d (SRK2I) after incubation. (C) Subcellular localization of Raf36-GFP in leaf mesophyll cells. Chl indicates chlorophyll autofluorescence. (Scale bar, 20 µm.) (D) BiFC assays for Raf36 and subclass III SnRK2s. SnRK2 and Raf36 or Raf43 were transiently expressed in N. benthamiana leaves by Agrobacterium infiltration. nEYFP and cEYFP represent the N- and C-terminal fragments of the EYFP, respectively. BF indicates bright field images. (Scale bar, 50 µm.) (E) Coimmunoprecipitation assay of Raf36 and SRK2I. Proteins were extracted from 35S:Raf36-HA and 35S:Raf36-HA/ 35S:FLAG-SRK2I plants treated with/without 50 µM ABA for 30 min. Immunoblotting (IB) was performed with an anti-HA or anti-FLAG antibody. (F) Y2H assay for truncated versions of Raf36 and SRK2E. Yeast cells coexpressing GAL4AD:Raf36, Raf36 N, or Raf36 KD+C and GAL4BD:SRK2E fusion proteins were incubated on SD media lacking L, W, H, and adenine (A), as follows (in order from low to high stringency): -LW, -LWH, -LWHA.
Next, we investigated which domain(s) of Raf36 may be responsible for the interaction with SnRK2s. According to the PROSITE database (https://prosite.expasy.org/), Raf36 contains an extended N-terminal stretch (N, 1 to 206 amino acids; aa) of unknown function, a predicted kinase catalytic domain (KD, 207 to 467 aa), and a short C-terminal domain (C, 468 to 525 aa) (Fig. 1F). In yeast two-hybrid assay, SRK2E strongly interacted with Raf36 full-length protein (FL) but showed slight or no interaction with Raf36 N and KD+C alone, respectively (Fig. 1F). These results indicated that the complete structure of Raf36 protein is required for interaction with SnRK2.
Raf36 Negatively Regulates ABA Response at Postgermination Growth Stage.
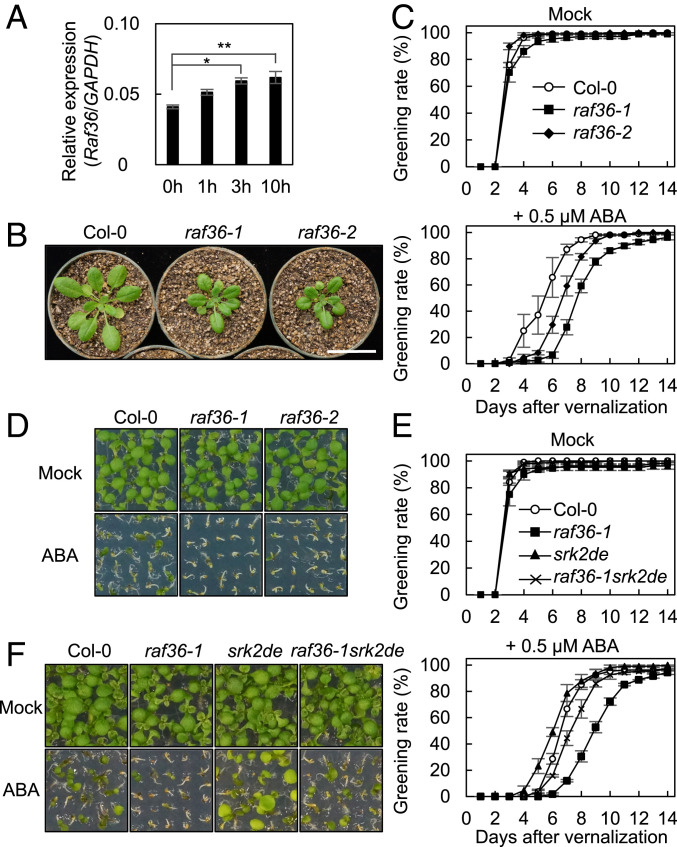
To characterize the role of Raf36 in ABA signaling, we performed a series of functional analyses in Arabidopsis. Using qRT-PCR, we measured the abundance of Raf36 messenger RNA (mRNA) in seedlings and detected a slight yet significant increase in Raf36 transcripts after ABA treatment (Fig. 2A). Next, we obtained two Raf36 transfer-DNA (T-DNA) insertion lines, GK-459C10 and SALK_044426C, designated as raf36-1 and raf36-2, respectively (SI Appendix, Fig. S4A). Using RT-PCR, we confirmed the loss of Raf36 transcripts in both mutants (SI Appendix, Fig. S4B). Notably, both of the raf36 mutants show growth retardation phenotypes under optimal growth conditions (Fig. 2B). We then measured rates of seed germination and cotyledon greening in the presence or absence of exogenous ABA. No difference in greening rate was observed between wild-type (WT) and mutant seedlings in the absence of ABA. However, with plants treated with 0.5 µM ABA, the greening rate of raf36 mutants was slower than WT (Fig. 2 C and D). This ABA-hypersensitive phenotype was complemented by CaMV35S:Raf36-GFP (SI Appendix, Fig. S4 C and D). To assess if the delayed greening of raf36 requires SnRK2 signaling, we generated a triple mutant, raf36-1srk2dsrk2e, and observed that it was less sensitive to ABA than raf36-1 (Fig. 2 E and F). This result indicates that SRK2D and SRK2E are genetic modifiers of raf36-dependent ABA hypersensitivity in the greening response. Notably, seed germination rates were not significantly changed in either raf36 mutant in the presence or absence of ABA (SI Appendix, Fig. S4E). Taken together, our results suggested that Raf36 functions as a negative regulator of SnRK2-dependent ABA signaling during postgerminative growth.
Fig. 2.
Raf36 negatively regulates SnRK2-dependent ABA response phenotypes in Arabidopsis seedlings. (A) Abundance of Raf36 mRNA transcripts measured by qRT-PCR. Total RNA was extracted from 1-wk-old WT Col-0 Arabidopsis seedlings treated with 50 µM ABA for indicated periods. Bars indicate means ± SE (n = 3), and asterisks showed significant differences by Dunnett’s test between ΔCt (nontreated) and ΔCt (ABA-treated) (*P < 0.05, **P < 0.01). (B) Dwarf phenotype of raf36 plants under normal condition. WT (Col-0), raf36-1, and raf36-2 plants grown at 22 °C under 16/8 h photoperiod for 29 d. (Scale bar, 3 cm.) (C and D) Quantification of the cotyledon greening rates of WT (Col-0), raf36-1, and raf36-2 on germination agar medium (GM) with or without 0.5 µM ABA. Data are means ± SE (n = 3). Each replicate contains 36 seeds. Photographs were taken 6 d after vernalization. (E and F) Quantification of the cotyledon greening rates of WT (Col-0), raf36-1, srk2dsrk2e, and raf36-1srk2dsrk2e on GM agar medium in the presence or absence of 0.5 µM ABA. Data are means ± SE (n = 3). Each replicate contains 36 seeds. Photographs were taken 6 d after vernalization.
Raf36 Is Phosphorylated by SnRK2.
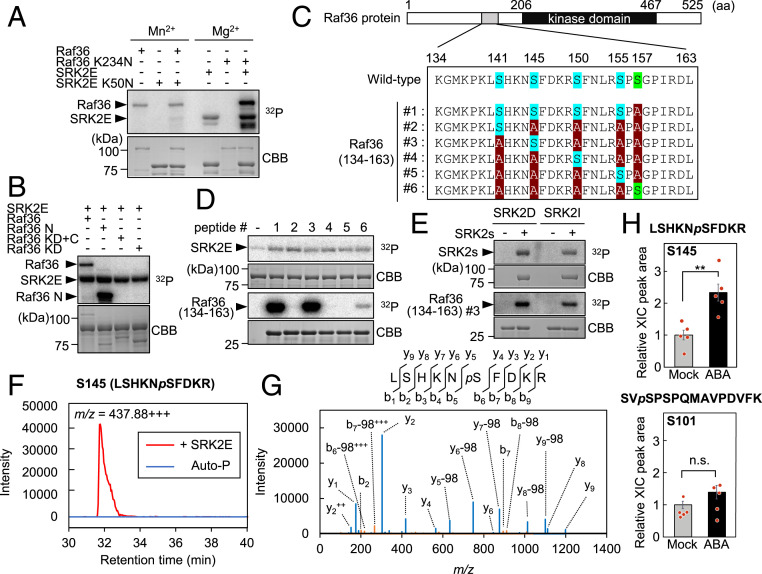
To further examine the biochemical relationship between SnRK2 and Raf36, we prepared Raf36 and SRK2E recombinant proteins as maltose-binding protein (MBP) or glutathione S-transferase (GST) fusions. Raf36 protein autophosphorylated, demonstrating that Raf36 is an active kinase (Fig. 3A). We found that Raf36 prefers Mn2+ for its kinase activity (SI Appendix, Fig. S5A), as shown for other C-group Raf (26–28), and SRK2E prefers Mg2+ for its kinase activity (SI Appendix, Fig. S5B). We then performed in vitro phosphorylation assays using kinase-dead forms of Raf36 and SRK2E as substrates. SRK2E phosphorylated Raf36 (K234N), while Raf36 did not phosphorylate SRK2E (K50N) (Fig. 3A). This result suggested that Raf36 is a potential substrate of SnRK2 but not vice versa.
Fig. 3.
Subclass III SnRK2s directly phosphorylate Raf36. (A) In vitro phosphorylation assay using kinase-dead forms of GST-SRK2E (SRK2E K50N) or MBP-Raf36 (Raf36 K234N). Each kinase-dead form was coincubated with an active GST-SRK2E or MBP-Raf36 kinase as indicated. Assays were performed in the presence of 5 mM Mn2+ (Left three lanes) or 5 mM Mg2+ (Right three lanes) with [γ-32P] ATP. (B) In vitro phosphorylation assay using truncated forms of MBP-tagged Raf36. Each MBP-Raf36 protein was incubated with MBP-SRK2E in the presence of 5 mM Mg2+ with [γ-32P] ATP. N: N-terminal region, KD: kinase domain, C: C-terminal region. (C) Schematic representation of six Raf36 (134 to 163) tested as SRK2E substrates. Ser141, Ser145, Ser150, and Ser155 are labeled in blue, with alanine substitutions shown in red. Ser157, labeled in green, was replaced with alanine in Raf36 (134 to 163) #1 to #5. (D) In vitro phosphorylation of GST-Raf36 (134-163) #1 to #6 by MBP-SRK2E. (E) In vitro phosphorylation of GST-Raf36 (134-163) #3 by MBP-SRK2D or MBP-SRK2I. Autoradiography (32P) and Coomassie brilliant blue (CBB) staining show protein phosphorylation and loading, respectively. (F and G) Phosphorylation of MBP-Raf36 Ser145 identified by phosphopeptide mapping. The amino acid sequences with probable phosphorylated serine residue (pS), ion-current chromatograms (F), and MS/MS spectrum (G) derived from a phosphopeptide of m/z = 437.88+++ are shown. MBP-Raf36 protein was either phosphorylated by MBP-SRK2E (+ SRK2E) or autophosphorylation (Auto-P) for 30 min. (H) Phosphorylation levels of Raf36 Ser145 (Upper) and Ser101 (Lower) after ABA treatment. Protein extracts were prepared from 2-wk-old Raf36pro:Raf36-3xFLAG/raf36-1 transgenic seedlings, which were treated with or without 50 µM ABA for 30 min, and total proteins were subjected to immunoprecipitation using anti-DYKDDDDK tag antibody beads followed by tryptic digestion. Peptides were analyzed by LC-MS/MS, and Raf36 phosphopeptides were identified and quantified based on the peak area of extracted ion chromatogram (XIC). Data are means ± SE from five independent biological replicates. Asterisks indicate significant differences (**P < 0.01, two-tailed Student’s t test). n.s., not significant.
Additional in vitro kinase assays were performed using a series of truncated versions of Raf36 to identify the phosphorylation site(s) of Raf36. First, we observed that Raf36 proteins lacking the N-terminal region (Raf 36 KD+C and Raf36 KD) were not phosphorylated, suggesting this region is important for both autophosphorylation by Raf36 and transphosphorylation by SRK2E (Fig. 3B). Second, Raf36 N (1 to 206) and Raf36 N (1 to 156) recombinant proteins were strongly phosphorylated, but Raf36 N (1 to 140) was only slightly phosphorylated by SRK2E (SI Appendix, Fig. S5C). These data suggested that the major phosphorylation site is located within 141 to 156 aa of the N terminus. Four serine (Ser) residues (i.e., Ser141, Ser145, Ser150, and Ser155) are present within this region. To identify which Ser residue(s) may be phosphorylated, we generated six peptides spanning this region of Raf36 (134 to 163). The peptides either contained all four serine residues (peptide #1) or had only a single Ser residue, with alanine substituted for the remaining Ser residues (peptides #2 to #6) (Fig. 3C). Ser157 was replaced with alanine in each peptide because it was outside of phosphorylated 141 to 156 aa region. Of these synthetic peptides, only Raf36 (134 to 163) #1 and #3 were strongly phosphorylated by SRK2E, indicating Ser145 is the phosphorylation site (Fig. 3D). SRK2D and SRK2I also phosphorylated Ser145 of Raf36 (134 to 163) in vitro (Fig. 3E). To test whether SnRK2 phosphorylates Ser145 of FL Raf36 protein, we conducted in vitro phosphorylation assay followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. The phosphopeptide containing Ser145 phosphorylation (LSHKNpSFDKR) was significantly enriched when MBP-Raf36 was reacted with MBP-SRK2E (Fig. 3 F and G). On the other hand, the peptide was barely detected when only MBP-Raf36 protein was used. In addition, this phosphopeptide was also found in vivo, and the abundance was significantly increased 2.3-fold in response to ABA, whereas the abundance of the other Raf36 N-terminal phosphopeptide containing phosphorylated Ser101 (SVpSPSPQMAVPDVFK) did not change (Fig. 3H). Together, these data strongly suggested that SnRK2s phosphorylate Raf36 at Ser145.
Raf22 Functions Partially Redundantly with Raf36.
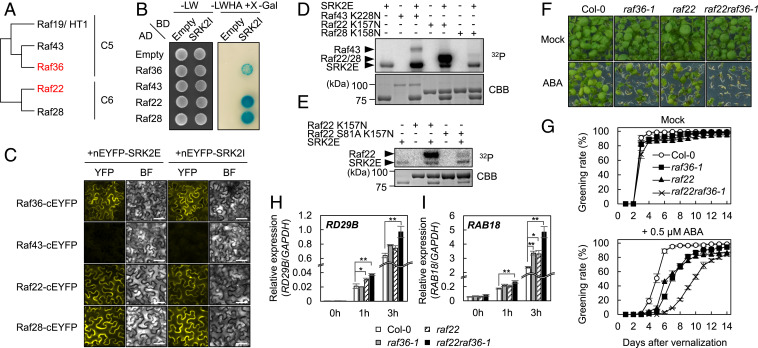
We next tested if Raf-like kinases closely related to Raf36 are also SnRK2 substrates. There are five kinases within Raf subgroups C5 and C6 (Fig. 4A). Among them, HIGH LEAF TEMPERATURE 1 (HT1/ Raf19) functions independently of ABA (29, 30). Therefore, we focused our analyses on Raf43, Raf22, and Raf28. As shown in Fig. 4B, SRK2I interacted with Raf36, Raf22, and Raf28, but not with Raf43, in yeast. Similarly, in BiFC assays, SRK2E and SRK2I interacted with Raf22 and Raf28 but not with Raf43 (Fig. 4C). To test whether SnRK2 phosphorylates these Raf kinases, in vitro phosphorylation assays were conducted using kinase-dead forms of Raf43 (Raf43 K228N), Raf22 (Raf22 K157N), or Raf28 (Raf28 K158N) as substrates. As shown in Fig. 4D, SRK2E strongly phosphorylated Raf22, despite having no equivalent of Ser145 of Raf36 and sharing only 29% identity with Raf36. In addition, SRK2E phosphorylated Raf43 but did not phosphorylate Raf28.
Fig. 4.
Raf22, a C6 Raf-like kinase, functions redundantly with Raf36. (A) Phylogenetic tree of subfamily C5 and C6 Raf-like kinases in Arabidopsis. (B) Y2H assay for C5/C6 Raf kinases and SRK2I. Yeast cells expressing GAL4AD:Raf and GAL4BD:SRK2I fusion proteins were grown on nonselective SD-LW (synthetic defined media lacking leucine and tryptophan) or selective SD-LWHA (SD lacking leucine, tryptophan, histidine, and adenine) media, and LacZ reporter activity was determined by X-Gal overlay assay. (C) BiFC assay for C5/C6 Raf kinases and SnRK2s (SRK2E and SRK2I) in N. benthamiana leaves. nEYFP and cEYFP represent the N- and C-terminal fragments of the EYFP, respectively. BF indicates bright field images. (Scale bar, 50 µm.) (D) In vitro phosphorylation of C5/C6 Raf kinases by GST-tagged SRK2E. MBP-tagged kinase-dead forms of Raf43 (Raf43 K228N), Raf22 (Raf22 K157N), or Raf28 (Raf28 K158N) were used as substrates. (E) In vitro phosphorylation of kinase-dead Raf22 (K157N) and Raf22 (S81A K157N) proteins by GST-SRK2E. (F and G) Quantification of the cotyledon greening rates of WT (Col-0), raf36-1, raf22, and raf22raf36-1 on GM agar medium with or without 0.5 µM ABA. Data are means ± SE (n = 4). Each replicate contains 36 seeds. Photographs were taken 9 d after vernalization. (H and I) Relative gene expression of ABA-responsive genes. Total RNA was extracted from 1-wk-old plants including WT, raf36-1, raf22, and raf22raf36-1 treated with 50 µM ABA for indicated periods. Bars indicate means ± SE (n = 3), and asterisks showed significant differences by Dunnett’s test between ΔCt (Col-0) and ΔCt (mutant) for each time point (*P < 0.05, **P < 0.01).
Because Raf22 did not share an equivalently conserved Ser145 as had been identified as the phosphorylation target in Raf36, we sought to identify the phosphorylation site targeted by SnRK2 in Raf22. As described above, Raf28 was not phosphorylated by SnRK2 (Fig. 4D), despite having 88% identity with Raf22. Because SnRK2 kinases prefer [-(R/K)-x-x-(S/T)-] or [-(S/T)-x-x-x-x-(E/D)-] (5, 7), we searched the amino acid sequences of Raf22 and Raf28 for potential SnRK2 phosphorylation sites and identified Ser81 within an [-R-H-Y-S-] motif in Raf22 that is converted to [-R-H-P-Y-S-] in Raf28. We introduced an alanine substitution at Ser81 in recombinant Raf22 and observed that this substitution nearly abolished phosphorylation by SRK2E (Fig. 4E), indicating this serine residue is an SnRK2-phosphorylation site in vitro. Collectively, we concluded that Raf36 and Raf22 are substrates of ABA-responsive SnRK2s.
Next, we functionally characterized the role of Raf22 in ABA-related phenotypes. Similar to Raf36, the transcriptional level of Raf22 was slightly up-regulated after exogenous ABA treatment (SI Appendix, Fig. S6A). A T-DNA insertional mutant (SALK_105195C), raf22, showed a similar phenotype to raf36-1, that is, ABA hypersensitivity in the postgermination growth (Fig. 4 F and G) but not in seed germination (SI Appendix, Fig. S6B).
To test potential functional redundancy between Raf36 and Raf22, a raf22raf36-1 double knockout mutant was generated. In the presence of ABA, raf22raf36-1 showed a stronger ABA-hypersensitive phenotype relative to individual raf22 and raf36-1 mutants (Fig. 4 F and G). In addition, expression of ABA- and stress-responsive genes RD29B and RAB18 were hyperinduced in raf22raf36-1 seedlings (Fig. 4 H and I). Because ABA also controls leaf water loss by inducing stomatal closure or other processes, we also measured water loss from detached leaves. In contrast to seedling greening phenotypes, leaf water loss of raf22raf36-1 and individual raf22 and raf36-1 plants was similar to that of WT plants (SI Appendix, Fig. S6C). In addition, leaf water loss of raf36-1srk2dsrk2e was similar to that of srk2dsrk2e plants (SI Appendix, Fig. S6C). Taken together, these results demonstrated that Raf36 and Raf22 function redundantly in ABA signaling during the postgerminative growth stage but are not required for ABA regulation of leaf water loss.
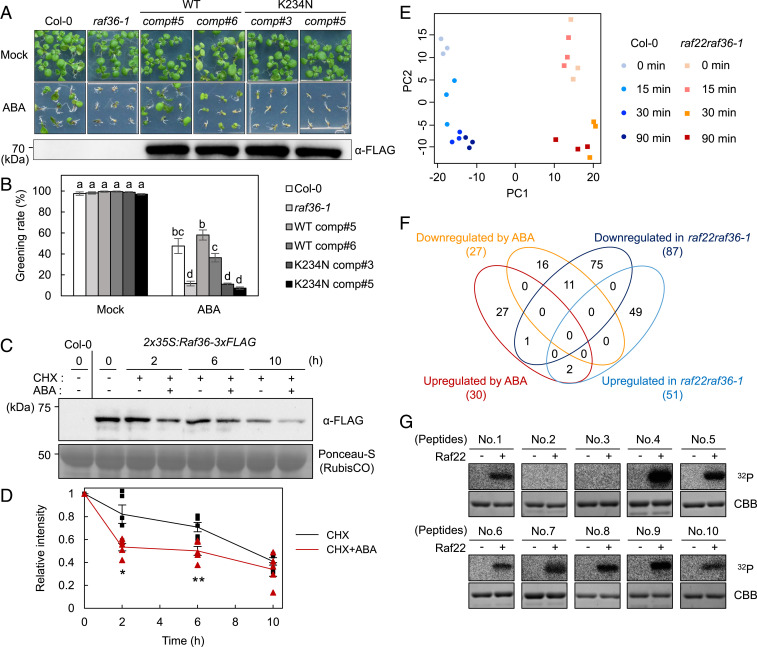
To examine whether the protein kinase activities of Raf36 and Raf22 are required for their negative regulation of ABA response, several complemented lines with kinase-dead form were generated. As shown in Fig. 5 A and B and SI Appendix, Fig. S7A, both the native promoter (Raf36pro)-driven and CaMV35S promoter-driven WT Raf36 complemented the ABA-hypersensitive phenotype of raf36-1, whereas the kinase-dead form of Raf36 (Raf36 K234N) could not. Similar results were also observed for Raf22 (SI Appendix, Fig. S7B). These results suggested that the protein kinase activities of Raf36 and Raf22 are required for their function in ABA signaling.
Fig. 5.
Phosphoproteomic analysis of WT and raf22raf36-1 identifies ABA signaling components downstream of Raf kinases. (A and B) Functional complementation of raf36-1 by Raf36pro:Raf36-3xFLAG or Raf36pro:Raf36 K234N-3xFLAG. Shown is photograph of seedlings grown for 7 d on GM agar medium in the presence or absence of 0.5 µM ABA. The Western blot image shows the expression level of Raf36-3xFLAG protein in 1-wk-old seedlings grown on normal GM agar medium. Greening rates were scored 7 d after vernalization. Data are means ± SE (n = 3), and each replicate contains 54 seeds. Different letters indicate significant differences (Tukey’s test, P < 0.01). (C and D) Western blot analysis of Raf36-3xFLAG after ABA treatment. Protein extracts were prepared from 2-wk-old WT (Col-0) or 2x35S:Raf36-3xFLAG transgenic plants, which were treated with 50 µM CHX in the presence or absence of 50 µM ABA for the indicated time periods. The Raf36 protein was detected by Western blotting using anti-FLAG antibody. Ponceau-S staining was used as loading control. Raf36 protein level at 0 h was set to 1.0 as a reference for calculating the relative band intensities at the various time points. The values are presented as the means ± SE from five independent biological replicates. Asterisks indicate significant differences for each time point (*P < 0.05, **P < 0.01, two-tailed Student’s t test). (E) PCA of quantitative data of phosphopeptides from WT (Col-0) and raf22raf36-1. (F) Venn diagram of up- or down-regulated phosphopeptides in WT seedlings after 50 µM ABA treatment, and up- or down-regulated phosphopeptides in raf22raf36-1 compared with WT. (G) In vitro phosphorylation of GST-tagged peptides from AT5G04740.1 (124 to 154 aa, No. 4), AT5G16260.1 (141 to 171 aa, No. 5), AT3G23920.1 (42 to 72 aa, No. 6), AT1G70770.1 (72 to 102 aa, No. 7), AT3G54610.1 (69 to 99 aa, No. 8), AT2G24050.1 (18 to 48 aa, No. 9), and AT1G37130.1 (48 to 78 aa, No. 10) by MBP-Raf22. GST-OLE1 fragment (AT4G25140.1, 143 to 173 aa, No. 1) was used as a positive control. GST-tagged peptides from AT3G62800.1 (60 to 90 aa, No. 2) and AT2G02070.1 (56 to 86 aa, No. 3) were included as negative controls. The autoradiography (32P) and Coomassie brilliant blue (CBB) staining show protein phosphorylation and loading, respectively.
ABA Promotes Raf36 Degradation.
Given that Raf36 and Raf22 are phosphorylated by ABA-responsive SnRK2s (Figs. 3 B–G and 4E), ABA might affect some aspects of Raf kinase function. To test for possible effects on Raf36 protein stability, we conducted immunoblotting using two transgenic plants (2x35S:Raf36-3xFLAG and 35S:Raf36-HA) after 50 µM ABA treatment for 0, 2, 6, and 10 h. In this assay, cycloheximide (CHX), a protein biosynthesis inhibitor, was used to eliminate the complication of newly synthesized Raf36. As shown in Fig. 5 C and D and SI Appendix, Fig. S8A, Raf36 protein level in plants treated with both CHX and ABA was significantly lower than that of the plants treated with only CHX. Intriguingly, ABA-mediated Raf36 degradation was alleviated when 35S:Raf36-HA plants were crossed with abi1-1C, a gain-of-function PP2C ABA-INSENSITIVE 1 (ABI1) mutant (14) (SI Appendix, Fig. S8B). These results suggest that Raf36 degradation is enhanced by ABA through PP2C-SnRK2 modules in planta. To test whether ABA can affect Raf kinase activity, we immunoprecipitated Raf36 (or Raf22) for kinase assays. However, we were unable to detect any Raf kinase activity in these experiments. Instead, in vitro phosphorylation assays demonstrated that Raf36 phospho-null (S145A) and phospho-mimetic (S145D) variants showed similar transphosphorylation activities on α-casein (SI Appendix, Fig. S9). Also, no remarkable change was observed even when introducing additional amino acid substitutions on Ser157, which was slightly phosphorylated by MBP-SRK2E in vitro (Fig. 3D and SI Appendix, Fig. S9). These results suggest that the N-terminal phosphorylation of Raf36 by SnRK2 may affect Raf36 protein stability rather than its kinase catalytic activity.
Phosphorylation Network Regulated by Raf22 and/or Raf36 Revealed by Comparative Phosphoproteomic Analysis.
To gain insight into the ABA-responsive signaling pathway(s) that may be regulated by Raf36 and/or Raf22, we performed a comparative phosphoproteomic analysis of WT and raf22raf36-1 seedlings treated with 50 µM ABA for 0, 15, 30, and 90 min. LC-MS/MS analysis identified a total of 2,093 phosphopeptides, and 1,066 of them were quantified in both WT and the raf22raf36-1 double mutant (Dataset S1). Within this dataset, 99.0% of identified phosphopeptides were singly phosphorylated (SI Appendix, Fig. S10A). Phosphoserine, phosphothreonine, and phosphotyrosine accounted for 91.0, 8.4, and 0.6% of phosphorylated residues, respectively (SI Appendix, Fig. S10B). Consistent with the ABA-hypersensitive phenotype observed in the raf22raf36-1 double mutant (Fig. 4 F and G), principal component analysis (PCA) demonstrated that the phosphoproteome profiles of the ABA-treated raf22raf36-1 mutant were different from that of ABA-treated WT (Fig. 5E). In addition, the profiles of raf22raf36-1 were significantly altered even before ABA treatment (Fig. 5E). This observation was consistent with our result in which Raf36 protein accumulated to a higher level under control conditions, as opposed to lower levels of Raf36 after ABA treatment (Fig. 5 C and D and SI Appendix, Fig. S8A).
Next, ABA-associated phosphopeptides differentially regulated between WT and raf22raf36-1 were identified. We selected ABA-responsive phosphopeptides with greater than 2-fold change (up-regulated by ABA) and with less than 1/2-fold change (down-regulated by ABA) with P < 0.05 (two-tailed Student’s t test) after ABA treatment for at least two of the time points in WT. Raf36- and/or Raf22-dependent phosphopeptides were selected with the following criteria: 1) a more than twofold increase (up-regulated in raf22raf36-1) or decrease (down-regulated in raf22raf36-1) in abundance compared with WT at each corresponding time point, and 2) the abundance of the phosphopeptide was significantly different from that of WT for at least two of the time points (P < 0.05; two-tailed Student’s t test). Based on these criteria, 30 phosphopeptides were up-regulated and 27 phosphopeptides were down-regulated by ABA treatment in WT plants (Fig. 5F and Dataset S2). Among the ABA up-regulated phosphopeptides, previously reported ABA-responsive phosphopeptides such as “EQS[+80]QVELELR” from SnRK2-substrate 1 (SNS1) (7), “QGS[+80]LTLPR” from abscisic acid responsive element-binding factors (ABFs) (8), and “S[+80]TVGTPAYIAPEVLLK” from SRK2E (SnRK2.6/OST1) (7) were identified, confirming the quality of the data in this study. In comparison with WT at each corresponding time point, a total of 51 and 87 phosphopeptides were up-regulated and down-regulated in raf22raf36-1, respectively (Fig. 5F and Dataset S2). Intriguingly, of the 27 phosphopeptides down-regulated by ABA in WT, 40.7% (11/27) were down-regulated in raf22raf36-1 mutant, indicating that Raf22 and/or Raf36 are mainly associated with ABA down-regulated phosphopeptides in planta.
Subsets of phosphopeptides were further analyzed to evaluate potential differences in cellular responses to ABA between WT and raf22raf36-1 plants. First, gene ontology (GO) analysis reported protein catabolism-related GO terms as well as the term “response to abscisic acid” as significantly up-regulated in the raf22raf36-1 double mutant (SI Appendix, Fig. S11C and Dataset S3), indicating that some ABA-responsive phosphopeptides were differentially regulated between WT and raf22raf36-1. In addition, terms such as “response to heat” and “innate immune response” were reported as significantly down-regulated in raf22raf36-1 (SI Appendix, Fig. S11D). Intriguingly, in raf22raf36-1, the term “abscisic acid-activated signaling pathway” was reported as enriched even before ABA treatment (SI Appendix, Fig. S12A). Second, we analyzed differentially accumulating phosphopeptides for enrichment of motifs that correspond to kinase recognition sequences. Analysis using rmotifx (31) identified two motifs, [-p(S/T)-P-] and [-R-x-x-p(S/T)-], that are MAPK- and SnRK2-/Calcium Dependent Protein Kinase (CDPK)-targeted sequences, respectively, as enriched in phosphopeptides from ABA-treated WT (SI Appendix, Fig. S13 A and B and Dataset S4). In addition, [-p(S/T)-P-] and [-R-x-x-p(S/T)-] motif containing phosphopeptides were also enriched in the up-regulated group in raf22raf36-1 (SI Appendix, Fig. S13C). Enriched motifs in the down-regulated group in raf22raf36-1 included [-p(S/T)-P-] along with [-p(S/T)-x-x-E-] (SI Appendix, Fig. S13D), suggesting that in planta Raf22 and/or Raf36 tend to prefer [-p(S/T)-P-] and [-p(S/T)-x-x-E-] motifs or alternatively that Raf22 and Raf36 regulate kinases that phosphorylate these motifs.
To test if some of the candidates from the phosphoproteomic data may be direct substrates of the Raf kinases, a subset of peptides was selected from the down-regulated group in raf22raf36-1. These candidates were synthesized as GST-fused, 31 amino acid–long peptides (peptide No. 4 to No.10) and used in an in vitro phosphorylation assay with recombinant Raf22. The in vitro phosphorylation assay demonstrated that Raf22 can phosphorylate AT5G04740.1: ACT DOMAIN REPEATS 12 (ACR12), AT5G16260.1: EARLY FLOWERING 9 (ELF9), AT3G23920.1: BETA-AMYLASE 1 (BAM1), AT1G70770.1: transmembrane protein, AT3G54610.1: HISTONE ACETYLTRANSFERASE 1 (HAT1, also known as GCN5 or HAG1), AT2G24050.1: EUKARYOTIC TRANSLATION INITIATION FACTOR ISOFORM 4G2 (eIFiso4G2), and AT1G37130.1: NITRATE REDUCTASE 2 (NIA2) (Fig. 5G). OLEOSIN1 (OLE1) was reported as a substrate of Raf22 in seeds (32) and was included as a positive control (peptide No. 1). In addition, peptides from proteins AT3G62800.1: DOUBLE-STRANDED-RNA-BINDING PROTEIN 4 (DRB4, peptide No.2) and AT2G02070.1: INDETERMINATE(ID)-DOMAIN 5 (IDD5, peptide No. 3) were selected from the Raf22/Raf36-independent group and included as negative controls. Together, these data indicate that at least a subset of phosphopeptides down-regulated in raf22raf36-1 are direct targets of Raf22 and/or Raf36.
Discussion
SnRK2s are core components of abiotic stress signaling in plants, yet the full complement of SnRK2 substrates leading to responses necessary for stress tolerance remain unknown. To gain a better understanding of SnRK2-mediated signaling pathway(s), we aimed to identify signaling factors associated with SnRK2. In this study, Raf36 and Raf22 were identified as SnRK2-interacting proteins. Using several reverse genetic and biochemical analyses, we revealed that Raf36 and Raf22 are SnRK2 substrates, which negatively regulate ABA responses in a partially redundant manner at the postgermination growth stage through their protein kinase activities. Inhibition of ABA signaling by these group C Raf-like kinases is likely to occur during optimal growth conditions, and ABA-activated SnRK2s induce Raf degradation to strengthen plant abiotic stress tolerance.
Physiological Functions of Raf22 and Raf36.
Our loss-of-function analyses revealed that Raf36 negatively regulates ABA responses (Fig. 2 C–F). Notably, loss of Raf36 altered the rate of cotyledon greening but did not affect seed germination or leaf water loss (SI Appendix, Fig. S4E and S6 B and C). These results suggest that Raf36 may function specifically in regulating ABA-induced postgermination growth arrest, a physiological response that may allow germinated seeds to survive under unfavorable conditions (25). Raf36 is a member of subgroup C5 Raf-like MAPKKK family in Arabidopsis. Previous studies also reported that other group C kinases, Raf22 and Raf43, are associated with ABA responses. In this regard, a raf22 mutant showed ABA-hypersensitive phenotype at the postgerminative growth stage (25) and raf43 mutant showed ABA-hypersensitive phenotype at the seed germination stage (24). Our results demonstrated that Raf36 and Raf22 function partially redundantly in ABA-mediated postgermination growth arrest, because raf22raf36-1 double knockout mutant showed a stronger ABA hypersensitivity than in the individual single mutants (Fig. 4 F and G). In addition to regulating postgermination growth arrest, our results show that Raf36 and Raf22 also regulate ABA-induced changes in gene expression and protein phosphorylation in older 1- or 2-wk-old seedlings (Figs. 4 H and I and 5 E and F, SI Appendix, Fig. S11 C and D). It is known that some group C Raf-like kinases function in various tissues, for example, HT1 or BLUE LIGHT-DEPENDENT H+-ATPASE PHOSPHORYLATION (BHP) in guard cells and Raf28 in embryogenesis (29, 33, 34). Thus, Raf36 and Raf22 may also have different functions in different tissues and developmental stages.
In our experiments, kinase-dead forms of Raf36 or Raf22 did not complement phenotypes of raf36-1 or raf22 mutants, indicating that in vivo kinase activity of both proteins is necessary for regulating ABA responses (Fig. 5 A and B and SI Appendix, Fig. S7 A and B). Phosphoproteomic analysis of ABA responses in WT and raf22raf36-1 double mutant plants revealed the phosphorylation network regulated by Raf36 and/or Raf22 and identified their possible substrates. Among phosphopeptides misregulated in raf22raf36-1, GO terms “response to abscisic acid” or “response to stress” were enriched (SI Appendix, Fig. S11 C and D), suggesting that Raf36 or Raf22 are involved in ABA-responsive phosphosignaling pathways. Consistent with this possibility, raf22raf36-1 double mutant showed enhanced salt tolerance compared with WT or the single mutant plants (SI Appendix, Fig. S14), indicating that Raf36 and Raf22 are involved in plant abiotic stress tolerance via ABA signaling. Importantly, the phosphoproteomic data strongly suggested that Raf36 and/or Raf22 function under optimal growth conditions, because PCA revealed phosphopeptide levels in raf22raf36-1 differed greatly from WT plants even before ABA treatment (Fig. 5E). Considering that the hyperinduction of ABA-responsive gene expression in raf22raf36-1 plants could be observed even at 1 h after ABA treatment (Fig. 4 H and I), the inhibition of ABA signaling by Raf36 and/or Raf22 seems to occur under optimal growth conditions or at initial stage of ABA responses. Consistent with this idea, immunoblot analysis demonstrated that Raf36 protein is more stable under control conditions than after ABA treatment (Fig. 5 C and D and SI Appendix, Fig. S8A). Furthermore, the slight growth retardation phenotypes of raf36, raf22raf36-1, or Raf36pro:Raf36 K234N-3xFLAG/raf36-1 plants supports a role for group C Raf-like kinase–mediated phosphosignaling in plant growth and development under optimal conditions (Fig. 2B and SI Appendix, Fig. S15). Interestingly, the GO term “innate immune response” was reported as down-regulated in raf22raf36-1 after ABA treatment (SI Appendix, Fig. S11D), indicating that Raf36 and/or Raf22 might also regulate defense signaling pathways, which are known to antagonize ABA signaling (35). Functional analyses of detected phosphoproteins will be required for further understanding of phosphosignaling pathways under the control of Raf36 or Raf22.
SnRK2 Regulates Raf36 and Raf22.
In this study, we identified Raf36 and Raf22 as direct targets of SnRK2s. In our data, SnRK2 directly phosphorylates the N-terminal region of Raf36 (Ser145) and Raf22 (Ser81) (Figs. 3 B–G and 4E). Recently, Lin et al. (21) analyzed a phosphoproteome of Arabidopsis WT plants and an SnRK2 decuple mutant. Their data demonstrated that Raf36 Ser145 and Raf22 Ser81 were phosphorylated in WT but not in a decuple mutant, suggesting that SnRK2s are responsible kinases for Raf22/36 phosphorylation in planta. SnRK2-mediated N-terminal phosphorylation of Raf36 may affect its protein stability because ABA treatment enhances Raf36 degradation in Arabidopsis (Fig. 5 C and D and SI Appendix, Fig. S8A) and because such ABA-mediated Raf36 degradation was alleviated in the plants harboring abi1-1 mutation, which inhibits ABA-responsive SnRK2 activation (14) (SI Appendix, Fig. S8B). Notably, although Raf36 (Ser145) does not correspond to the canonical SnRK2 consensus motifs such as [-(R/K)-x-x-(S/T)-] or [-(S/T)-x-x-x-x-(E/D)-] (5, 7), it corresponds to [-L-x-x-x-x-(S/T)-], which was recently reported as a new consensus motif for SRK2E (36). Evidence from other Raf-like kinases indicate that the N-terminal regions have roles in their molecular functions. CONSTITUTIVE TRIPLE RESPONSE 1 (CTR1/Raf1), a group B3 Raf, is a central negative regulator of ethylene signaling, and its N-terminal region interacts with a receptor ETHYLENE RESPONSE 1 (ETR1) (37–39). CONVERGENCE OF BLUE LIGHT AND CO2 1 (CBC1/Raf38), a group C7 Raf, regulates blue light- and low CO2–inducible stomatal opening, and its N-terminal region is phosphorylated by phototropin (40). In plant Raf-like protein kinases, KD are relatively conserved, but the N-terminal regions are more variable (15). Consistent with this, the SnRK2-phosphorylation sites of Raf36 (Ser145) and Raf22 (Ser81) were not identical. Therefore, the role of N-terminal regions may differ between Raf36 and Raf22. Further analysis will be required to understand the difference between the N-terminal phosphorylation of Raf36 and Raf22.
Previous studies have proposed the relationship between SnRK2 and Raf-like kinases. For instance, HT1/AtRaf19, a subgroup C5 Raf, regulates stomatal CO2 signaling with SRK2E/OST1/SnRK2.6 (41–43). Also, PpARK, a subgroup B3 Raf, was identified as a direct upstream regulator of SnRK2s that can activate SnRK2s by phosphorylating the activation loop of SnRK2 (16). Several recent studies reported that Arabidopsis B2, B3, and B4 subgroup of Raf-like kinases act as positive regulators upstream of SnRK2s in osmotic stress signaling (19–23). In contrast, our results demonstrate that group C Rafs negatively regulate ABA signaling under optimal growth conditions. Under unfavorable conditions, ABA-SnRK2 module induces Raf36 degradation, leading to a stronger ABA response for plants to adapt to such adverse environments. Understanding the interplay between group B and group C Rafs in regulating SnRK2 activity will be an important area of future research. Also, functional analyses of substrate candidates of Raf36 or Raf22, such as those identified in our phosphoproteomic screen, will increase understanding of how group C Rafs regulate ABA signaling.
Materials and Methods
Arabidopsis thaliana ecotype Columbia (Col-0) was used as the WT. T-DNA insertion mutant lines, raf36-1 (GK-459C10), raf36-2 (SALK_044426C), and raf22 (SALK_105195C) were obtained from Arabidopsis Biological Resource Center (ABRC) or GABI-Kat. The raf36-1raf22 double knockout mutant was generated by crossing raf36-1 and raf22. The SRK2E/OST1 knockout mutant, srk2e (SALK_008068), was used as described previously (10). The srk2dsrk2e double mutant was established by crossing srk2d (GABI-Kat 807G04) and srk2e, and the raf36-1srk2dsrk2e triple mutant was generated by crossing srk2dsrk2e double mutant and raf36-1. Detailed information on the plant growth conditions was described in SI Appendix. In addition, details of plasmid constructions, protein–protein interaction assays, microscopy analyses of fluorescent proteins, preparation of recombinant proteins, in vitro phosphorylation assays, water loss analysis, RNA extraction and qRT-PCR, and phosphoproteomic analysis were provided in SI Appendix, Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Tsuyoshi Nakagawa (Shimane University, Japan) for providing the R4pGWB501 vector and Dr. Yoichi Sakata (Tokyo University of Agriculture, Japan), Dr. José M. Barrero (Commonwealth Scientific and Industrial Research Organization; CSIRO, Australia), and Dr. Paul E. Verslues (Academia Sinica, Taiwan) for valuable discussion. We also thank Tomotaka Itaya, Ryo Yoshimura (Nagoya University, Japan), and Mrs. Saho Mizukado (RIKEN, Japan) for their expert technical assistance. We are grateful to the ABRC and GABI-Kat project for providing Arabidopsis T-DNA insertional mutants. This work was partly supported by the Japan Society for the Promotion of Science KAKENHI Grants JP21J10962 to Y. Kamiyama, JP15H04383, 16KK0160, and 19H03240 to T.U., and JST PRESTO P13413773 to T.U.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2100073118/-/DCSupplemental.
Data Availability
LC-MS raw data and R file data have been deposited in Japan Proteome Standard Repository/Database (jPOST) and GitHub, respectively (https://repository.jpostdb.org/entry/JPST000961 [for jPOST]; and http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD021372 [for ProteomeXchange] and https://github.com/Ume-lab/raf22raf36_phosphoproteome [for GitHub]).
References
- 1.Finkelstein R., Abscisic acid synthesis and response. Arabidopsis Book 11, e0166 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shinozaki K., Yamaguchi-Shinozaki K., Seki M., Regulatory network of gene expression in the drought and cold stress responses. Curr. Opin. Plant Biol. 6, 410–417 (2003). [DOI] [PubMed] [Google Scholar]
- 3.Cutler S. R., Rodriguez P. L., Finkelstein R. R., Abrams S. R., Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 61, 651–679 (2010). [DOI] [PubMed] [Google Scholar]
- 4.Umezawa T., et al., Molecular basis of the core regulatory network in ABA responses: Sensing, signaling and transport. Plant Cell Physiol. 51, 1821–1839 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Furihata T., et al., Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U.S.A. 103, 1988–1993 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Geiger D., et al., Activity of guard cell anion channel SLAC1 is controlled by drought-stress signaling kinase-phosphatase pair. Proc. Natl. Acad. Sci. U.S.A. 106, 21425–21430 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Umezawa T., et al., Genetics and phosphoproteomics reveal a protein phosphorylation network in the abscisic acid signaling pathway in Arabidopsis thaliana. Sci. Signal. 6, rs8 (2013). [DOI] [PubMed] [Google Scholar]
- 8.Wang P., et al., Quantitative phosphoproteomics identifies SnRK2 protein kinase substrates and reveals the effectors of abscisic acid action. Proc. Natl. Acad. Sci. U.S.A. 110, 11205–11210 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hrabak E. M., et al., The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132, 666–680 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yoshida R., et al., ABA-activated SnRK2 protein kinase is required for dehydration stress signaling in Arabidopsis. Plant Cell Physiol. 43, 1473–1483 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Fujii H., Zhu J.-K., Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U.S.A. 106, 8380–8385 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fujita Y., et al., Three SnRK2 protein kinases are the main positive regulators of abscisic acid signaling in response to water stress in Arabidopsis. Plant Cell Physiol. 50, 2123–2132 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Nakashima K., et al., Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50, 1345–1363 (2009). [DOI] [PubMed] [Google Scholar]
- 14.Umezawa T., et al., Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 106, 17588–17593 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ichimura K.et al.; MAPK Group , Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 7, 301–308 (2002). [DOI] [PubMed] [Google Scholar]
- 16.Saruhashi M., et al., Plant Raf-like kinase integrates abscisic acid and hyperosmotic stress signaling upstream of SNF1-related protein kinase2. Proc. Natl. Acad. Sci. U.S.A. 112, E6388–E6396 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stevenson S. R., et al., Genetic analysis of Physcomitrella patens identifies ABSCISIC ACID NON-RESPONSIVE (ANR), a regulator of ABA responses unique to basal land plants and required for desiccation tolerance. Plant Cell 28, 1310–1327 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yasumura Y., et al., An ancestral role for CONSTITUTIVE TRIPLE RESPONSE1 proteins in both ethylene and abscisic acid signaling. Plant Physiol. 169, 283–298 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Katsuta S., et al., Arabidopsis Raf-like kinases act as positive regulators of subclass III SnRK2 in osmostress signaling. Plant J. 103, 634–644 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Takahashi Y., et al., MAP3Kinase-dependent SnRK2-kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat. Commun. 11, 12 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Z., et al., A RAF-SnRK2 kinase cascade mediates early osmotic stress signaling in higher plants. Nat. Commun. 11, 613 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nguyen Q. T. C., et al., Arabidopsis Raf-like kinase Raf10 is a regulatory component of core ABA signaling. Mol. Cells 42, 646–660 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soma F., Takahashi F., Suzuki T., Shinozaki K., Yamaguchi-Shinozaki K., Plant Raf-like kinases regulate the mRNA population upstream of ABA-unresponsive SnRK2 kinases under drought stress. Nat. Commun. 11, 1373 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Virk N., et al., Arabidopsis Raf-like mitogen-activated protein kinase kinase kinase gene Raf43 is required for tolerance to multiple abiotic stresses. PLoS One 10, e0133975 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hwang J.-U., Yim S., Do T. H. T., Kang J., Lee Y., Arabidopsis thaliana Raf22 protein kinase maintains growth capacity during postgerminative growth arrest under stress. Plant Cell Environ. 41, 1565–1578 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Lamberti G., Gügel I. L., Meurer J., Soll J., Schwenkert S., The cytosolic kinases STY8, STY17, and STY46 are involved in chloroplast differentiation in Arabidopsis. Plant Physiol. 157, 70–85 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reddy M. M., Rajasekharan R., Role of threonine residues in the regulation of manganese-dependent Arabidopsis serine/threonine/tyrosine protein kinase activity. Arch. Biochem. Biophys. 455, 99–109 (2006). [DOI] [PubMed] [Google Scholar]
- 28.Rudrabhatla P., Reddy M. M., Rajasekharan R., Genome-wide analysis and experimentation of plant serine/ threonine/tyrosine-specific protein kinases. Plant Mol. Biol. 60, 293–319 (2006). [DOI] [PubMed] [Google Scholar]
- 29.Hashimoto M., et al., Arabidopsis HT1 kinase controls stomatal movements in response to CO2. Nat. Cell Biol. 8, 391–397 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Hashimoto-Sugimoto M., et al., Dominant and recessive mutations in the Raf-like kinase HT1 gene completely disrupt stomatal responses to CO2 in Arabidopsis. J. Exp. Bot. 67, 3251–3261 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wagih O., Sugiyama N., Ishihama Y., Beltrao P., Uncovering phosphorylation-based specificities through functional interaction networks. Mol. Cell. Proteomics 15, 236–245 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ramachandiran I., Vijayakumar A., Ramya V., Rajasekharan R., Arabidopsis serine/threonine/tyrosine protein kinase phosphorylates oil body proteins that regulate oil content in the seeds. Sci. Rep. 8, 1154 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hayashi M., Inoue S. I., Ueno Y., Kinoshita T., A Raf-like protein kinase BHP mediates blue light-dependent stomatal opening. Sci. Rep. 7, 45586 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang B., et al., The RAF-like mitogen-activated protein kinase kinase kinases RAF22 and RAF28 are required for the regulation of embryogenesis in Arabidopsis. Plant J. 96, 734–747 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Ton J., Flors V., Mauch-Mani B., The multifaceted role of ABA in disease resistance. Trends Plant Sci. 14, 310–317 (2009). [DOI] [PubMed] [Google Scholar]
- 36.Wang P., et al., Mapping proteome-wide targets of protein kinases in plant stress responses. Proc. Natl. Acad. Sci. U.S.A. 117, 3270–3280 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Clark K. L., Larsen P. B., Wang X., Chang C., Association of the Arabidopsis CTR1 Raf-like kinase with the ETR1 and ERS ethylene receptors. Proc. Natl. Acad. Sci. U.S.A. 95, 5401–5406 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang Y., Li H., Hutchison C. E., Laskey J., Kieber J. J., Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J. 33, 221–233 (2003). [DOI] [PubMed] [Google Scholar]
- 39.Kieber J. J., Rothenberg M., Roman G., Feldmann K. A., Ecker J. R., CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the Raf family of protein kinases. Cell 72, 427–441 (1993). [DOI] [PubMed] [Google Scholar]
- 40.Hiyama A., et al., Blue light and CO2 signals converge to regulate light-induced stomatal opening. Nat. Commun. 8, 1284 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hõrak H., et al., A dominant mutation in the HT1 kinase uncovers roles of MAP kinases and GHR1 in CO2-induced stomatal closure. Plant Cell 28, 2493–2509 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tian W., et al., A molecular pathway for CO2 response in Arabidopsis guard cells. Nat. Commun. 6, 6057 (2015). [DOI] [PubMed] [Google Scholar]
- 43.Matrosova A., et al., The HT1 protein kinase is essential for red light-induced stomatal opening and genetically interacts with OST1 in red light and CO2 -induced stomatal movement responses. New Phytol. 208, 1126–1137 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
LC-MS raw data and R file data have been deposited in Japan Proteome Standard Repository/Database (jPOST) and GitHub, respectively (https://repository.jpostdb.org/entry/JPST000961 [for jPOST]; and http://proteomecentral.proteomexchange.org/cgi/GetDataset?ID=PXD021372 [for ProteomeXchange] and https://github.com/Ume-lab/raf22raf36_phosphoproteome [for GitHub]).