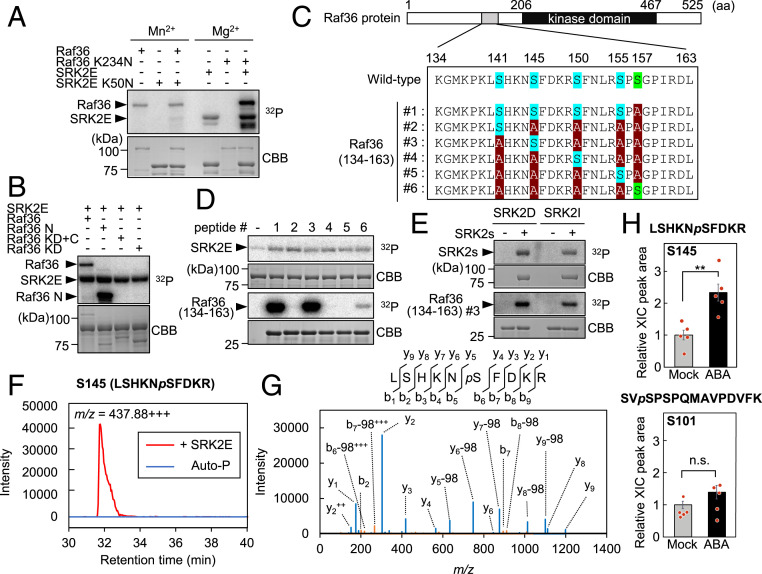
Fig. 3.
Subclass III SnRK2s directly phosphorylate Raf36. (A) In vitro phosphorylation assay using kinase-dead forms of GST-SRK2E (SRK2E K50N) or MBP-Raf36 (Raf36 K234N). Each kinase-dead form was coincubated with an active GST-SRK2E or MBP-Raf36 kinase as indicated. Assays were performed in the presence of 5 mM Mn2+ (Left three lanes) or 5 mM Mg2+ (Right three lanes) with [γ-32P] ATP. (B) In vitro phosphorylation assay using truncated forms of MBP-tagged Raf36. Each MBP-Raf36 protein was incubated with MBP-SRK2E in the presence of 5 mM Mg2+ with [γ-32P] ATP. N: N-terminal region, KD: kinase domain, C: C-terminal region. (C) Schematic representation of six Raf36 (134 to 163) tested as SRK2E substrates. Ser141, Ser145, Ser150, and Ser155 are labeled in blue, with alanine substitutions shown in red. Ser157, labeled in green, was replaced with alanine in Raf36 (134 to 163) #1 to #5. (D) In vitro phosphorylation of GST-Raf36 (134-163) #1 to #6 by MBP-SRK2E. (E) In vitro phosphorylation of GST-Raf36 (134-163) #3 by MBP-SRK2D or MBP-SRK2I. Autoradiography (32P) and Coomassie brilliant blue (CBB) staining show protein phosphorylation and loading, respectively. (F and G) Phosphorylation of MBP-Raf36 Ser145 identified by phosphopeptide mapping. The amino acid sequences with probable phosphorylated serine residue (pS), ion-current chromatograms (F), and MS/MS spectrum (G) derived from a phosphopeptide of m/z = 437.88+++ are shown. MBP-Raf36 protein was either phosphorylated by MBP-SRK2E (+ SRK2E) or autophosphorylation (Auto-P) for 30 min. (H) Phosphorylation levels of Raf36 Ser145 (Upper) and Ser101 (Lower) after ABA treatment. Protein extracts were prepared from 2-wk-old Raf36pro:Raf36-3xFLAG/raf36-1 transgenic seedlings, which were treated with or without 50 µM ABA for 30 min, and total proteins were subjected to immunoprecipitation using anti-DYKDDDDK tag antibody beads followed by tryptic digestion. Peptides were analyzed by LC-MS/MS, and Raf36 phosphopeptides were identified and quantified based on the peak area of extracted ion chromatogram (XIC). Data are means ± SE from five independent biological replicates. Asterisks indicate significant differences (**P < 0.01, two-tailed Student’s t test). n.s., not significant.