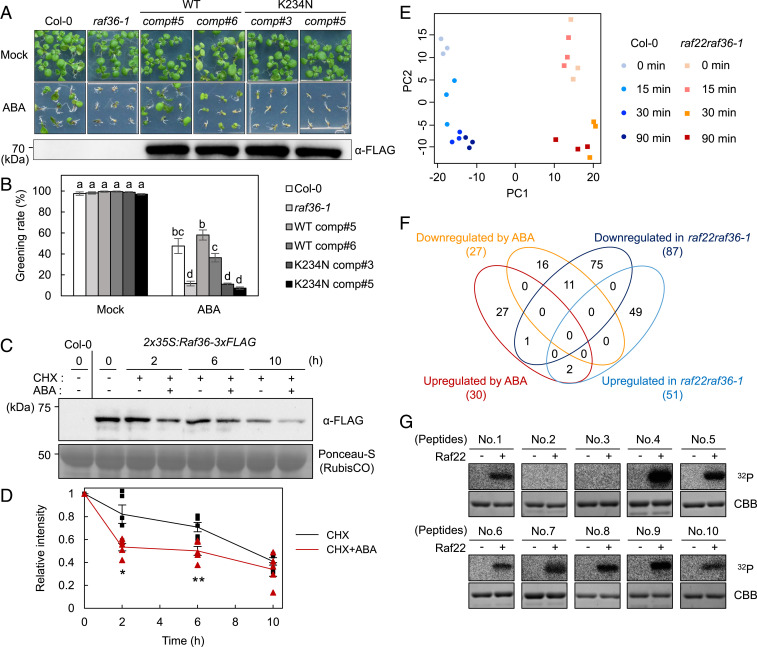
Fig. 5.
Phosphoproteomic analysis of WT and raf22raf36-1 identifies ABA signaling components downstream of Raf kinases. (A and B) Functional complementation of raf36-1 by Raf36pro:Raf36-3xFLAG or Raf36pro:Raf36 K234N-3xFLAG. Shown is photograph of seedlings grown for 7 d on GM agar medium in the presence or absence of 0.5 µM ABA. The Western blot image shows the expression level of Raf36-3xFLAG protein in 1-wk-old seedlings grown on normal GM agar medium. Greening rates were scored 7 d after vernalization. Data are means ± SE (n = 3), and each replicate contains 54 seeds. Different letters indicate significant differences (Tukey’s test, P < 0.01). (C and D) Western blot analysis of Raf36-3xFLAG after ABA treatment. Protein extracts were prepared from 2-wk-old WT (Col-0) or 2x35S:Raf36-3xFLAG transgenic plants, which were treated with 50 µM CHX in the presence or absence of 50 µM ABA for the indicated time periods. The Raf36 protein was detected by Western blotting using anti-FLAG antibody. Ponceau-S staining was used as loading control. Raf36 protein level at 0 h was set to 1.0 as a reference for calculating the relative band intensities at the various time points. The values are presented as the means ± SE from five independent biological replicates. Asterisks indicate significant differences for each time point (*P < 0.05, **P < 0.01, two-tailed Student’s t test). (E) PCA of quantitative data of phosphopeptides from WT (Col-0) and raf22raf36-1. (F) Venn diagram of up- or down-regulated phosphopeptides in WT seedlings after 50 µM ABA treatment, and up- or down-regulated phosphopeptides in raf22raf36-1 compared with WT. (G) In vitro phosphorylation of GST-tagged peptides from AT5G04740.1 (124 to 154 aa, No. 4), AT5G16260.1 (141 to 171 aa, No. 5), AT3G23920.1 (42 to 72 aa, No. 6), AT1G70770.1 (72 to 102 aa, No. 7), AT3G54610.1 (69 to 99 aa, No. 8), AT2G24050.1 (18 to 48 aa, No. 9), and AT1G37130.1 (48 to 78 aa, No. 10) by MBP-Raf22. GST-OLE1 fragment (AT4G25140.1, 143 to 173 aa, No. 1) was used as a positive control. GST-tagged peptides from AT3G62800.1 (60 to 90 aa, No. 2) and AT2G02070.1 (56 to 86 aa, No. 3) were included as negative controls. The autoradiography (32P) and Coomassie brilliant blue (CBB) staining show protein phosphorylation and loading, respectively.