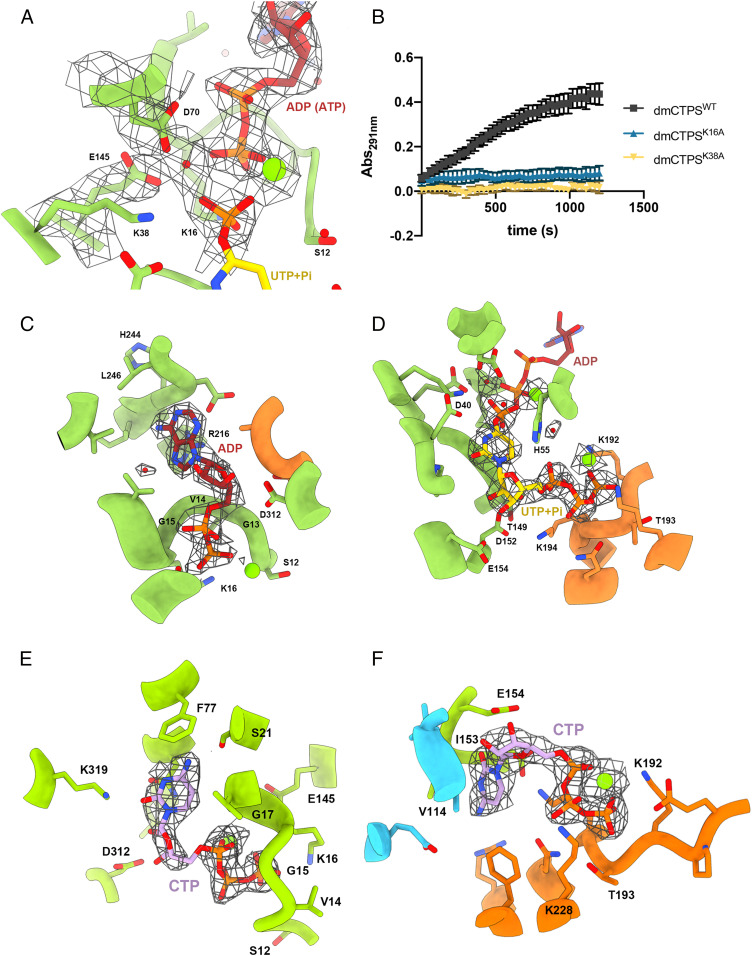
Fig. 5.
Ligands binding at AL domain reveal mechanisms of UTP phosphorylation and feedback inhibition. (A) Electron density map showing the mechanism of UTP phosphorylation. (B) Analysis for generation of CTP by K16A and K38A mutant dmCTPS in condition with 0.2 mM GTP. The absorption of 291 nm (Abs291nm) represents the concentration of CTP in samples. For the analysis, 2 μM dmCTPS was mixed with 2 mM UTP, 2 mM ATP, 10 mM MgCl2, 25 mM Tris HCl (pH 7.5), and 0.2 mM GTP. Glutamine (10 mM) is supplied for initiating the reaction. (Error bars, SD.) (C and D) The ATP (C) and UTP (D) binding site at AL domain. ATP and UTP have become ADP and phosphorylated UTP in our dmCTPS+Sub model. Residues interacting with ADP/UTP are indicated. (E and F) The competitive binding of CTP with ATP (E) and UTP (F). Water molecules are shown as red spheres in all panels.