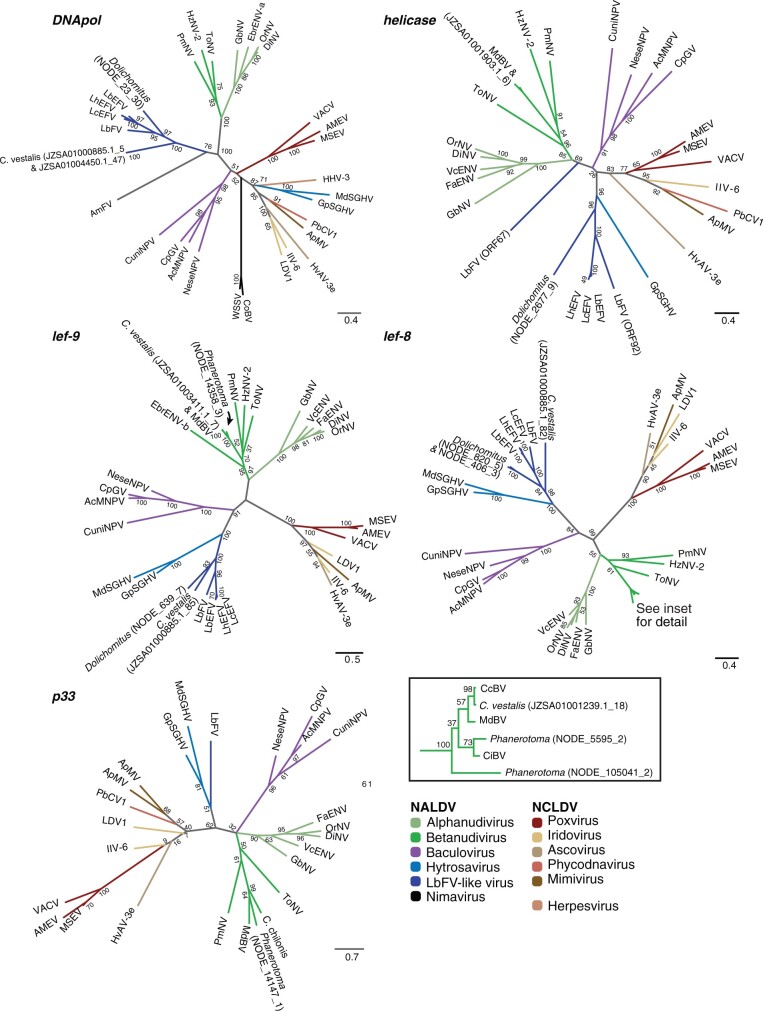
Fig. 2.
Genes of viral origin in wasp genomes are related to genes in large dsDNA viruses. Maximum likelihood phylogenies constructed from protein sequences of individual genes are shown, with numbers on nodes indicating support from 100 bootstrap replicates. Colored branches indicate virus families or groups. The sequences used for alignment were obtained from Microplitis demoltior bracovirus (MdBV), Cotesia congregata bracovirus (CcBV), Chelonus insularis bracovirus (CiBV), a C. chilonis transcriptome, Eurytoma brunniventris endogenous nudivirus alpha (EbENV-a), Eurytoma brunniventris endogenous nudivirus beta (EbENV-b), Tipula oleracea nudivirus (ToNV), Heliothis zea nudivirus 2 (HzNV-2), Penaeus monodon nudivirus (PmNV), Venturia canescens endogenous nudivirus (VcENV), Drosophila innubila nudivirus (DiNV), Oryctes rhinoceros nudivirus (OrNV), Fopius arisanus endogenous nudivirus (FaENV), Gryllus bimaculatus nudivirus (GbNV), Autographa californica multiple nucleopolyhedrovirus (AcMNPV), Cydia pomonella granulovirus (CpGV), Neodiprion sertifer nucleopolyhedrovirus (NeseMNPV), Culex nigripalpus nucleopolyhedrosis virus (CuniNPV), Glossina pallidipes salivary gland hytrosavirus (GpSGHV), and Musca domestica salivary gland hytrosavirus (MdSGHV), L. boulardi filamentous virus (LbFV), L. boulardi endogenous filamentous virus (LbEFV), Leptopilina heterotoma endogenous filamentous virus (LhEFV), Leptopilina clavipes endogenous filamentous virus (LcEFV), A. mellifera filamentous virus (AmFV), White spot syndrome virus (WSSV), Chionoecetes opilio bacilliform virus (CoBV), Amsacta moorei entomopoxvirus (AMEV), Melanoplus sanguinipes entomopoxvirus (MSEV), Vaccinia virus (VACV), Invertebrate iridescent virus 6 (IIV-6), LDV1, Trichoplusia ni ascovirus 3e (TnAV-3e), Paramecium bursaria chlorella virus 1 (PbCV1), Acanthamoeba polyphaga mimivirus (ApMV), Human herpesvirus 3 (HHV-3), supplementary table S5, Supplementary Material online. Sequences from the IVSPERs of the ichnovirus in Glypta fumiferanae and nimavirus p33 homologs were not included because they were too divergent compared with the other included sequences. Scale bars indicate substitutions per amino acid residue. For ORFs found in wasp genomes, the accession number or scaffold name is followed by an underscore and the ORF number.