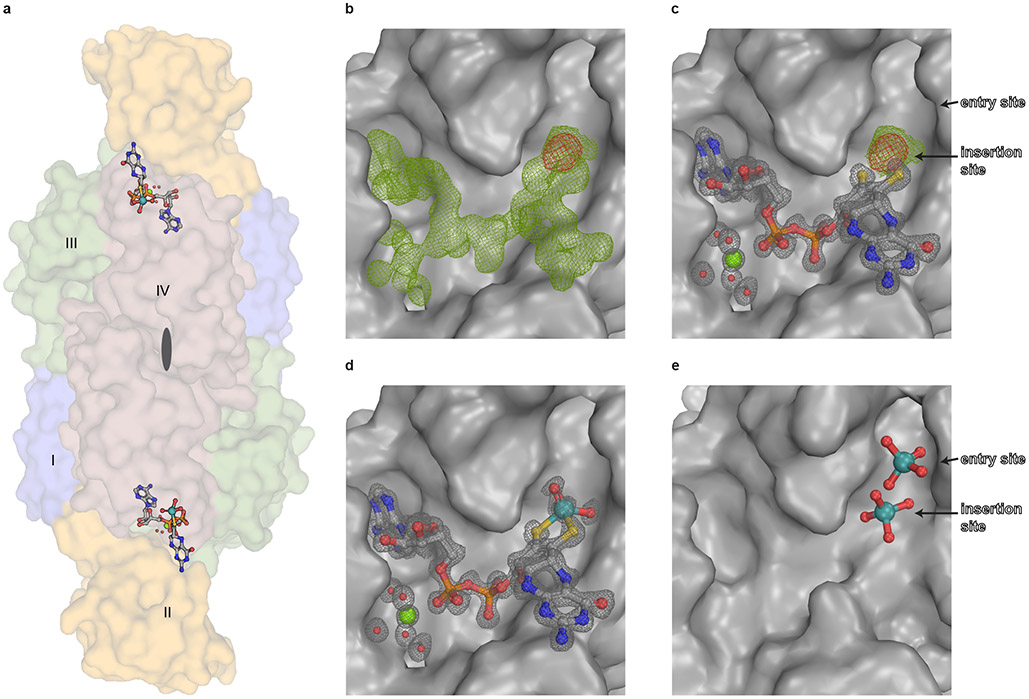
Figure 2: Structure of Cnx1E variant S269D D274S with bound ligands.
(A) Side view of the Cnx1E physiological dimer in Conolly-surface representation and transparent. The Cnx1E subdomains are colored as follows: subdomain I = slate-blue, subdomain II = yellow-orange, subdomain III = forest green, subdomain IV = dark salmon. Subdomains are indicated (I-IV) for monomer A. (B-E) Electron density of Cnx1E bound ligands. (B) Composite-omit map at 1.5 σ (green) and anomalous map at 3.5 σ (red) of the electron density found in the active site of Cnx1E variant S269D D274S. (C) Feature-enhanced map at 1.5 σ (gray) calculated in the presence of MPT-AMP. Composite map (green) and anomalous map (red) indicate the presence a molybdenum atom near the dithiolene motif. (D) Feature-enhanced map at 1.5 σ calculated in the presence of modeled Moco-AMP. (E) Oxo-anion binding pocket of Cnx1E as seen in the wildtype structure (PDB: 6ETF)29. (A-E) Ligands are shown in ball and stick representation or spheres (magnesium water complex). The following color code was applied: light gray, carbon; red, oxygen; blue, nitrogen; orange, phosphorous; yellow, sulfur; green, magnesium; teal, molybdenum.