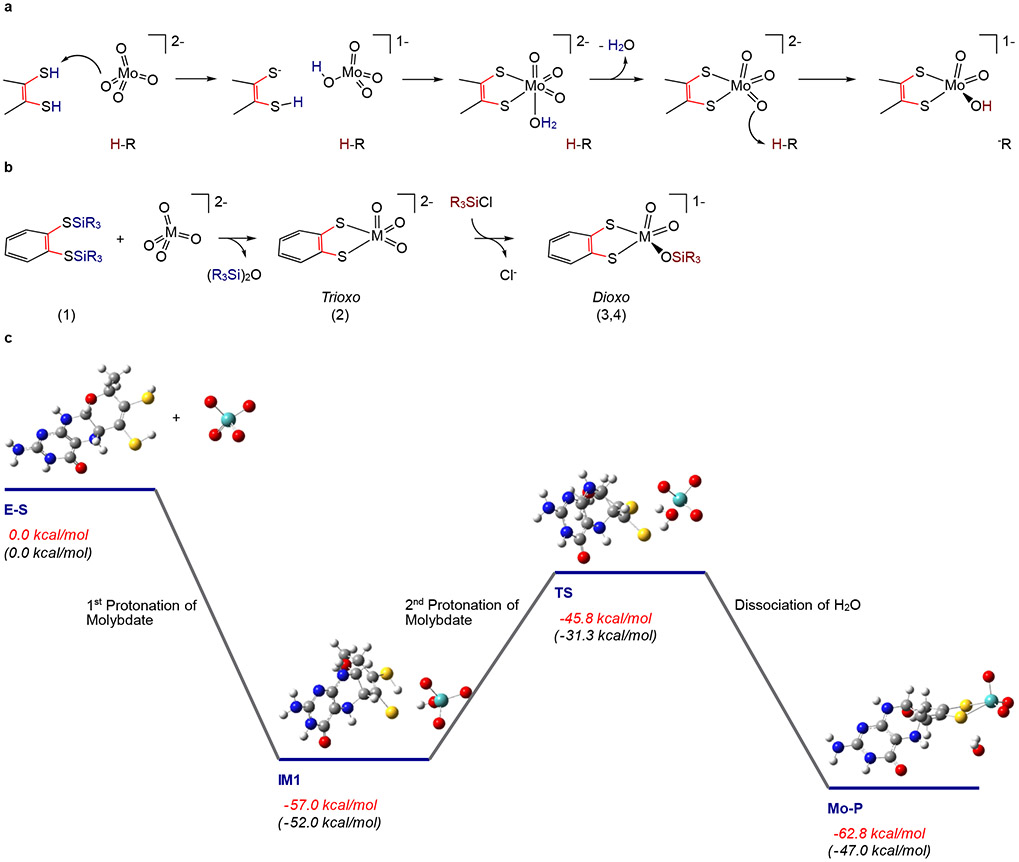
Figure 5: Proposed reaction mechanism for Cnx1E catalyzed Mo insertion.
(A) The mechanism of molybdate insertion consistent with X-ray crystallographic and XANES/EXAFS studies on Cnx1E variant S269D D274S. (B) Synthetic scheme for the synthesis of trioxo-(Mo/W) and dioxo-W small molecule analogs of the Cnx1E Mo site showing a remarkable similarity to the proposed mechanism for molybdate insertion catalyzed by Cnx1E. Note that in the small molecule synthetic scheme [SiR3]+ is the functional equivalent of the proton (H+) in the enzyme mechanism. (C) Computed reaction coordinate for the formation of the molybdenum cofactor that is consistent with the mechanism described in A. (E-S) A doubly protonated molybdopterin (MPT) is the likely candidate for activating one of the molybdate Mo-oxo bonds to eliminate water. Although the first protonation is barrierless (IM 1), the second protonation step (IM1→TS→Mo-P) to eliminate water and bind Mo to MPT (yielding [(MPT)MoO3]2−) occurs with a moderate activation barrier. The XAS results show that the initial [(MPT)MoO3]2−product must be further protonated at one of the oxo ligands to yield the [(MPT)MoO2(OH)]1− structure identified in Cnx1E bound Moco-AMP. Formation of [(MPT)MoO2(OH)]1− is expected to prevent the potential back reaction of Mo-P to IM1. Energies in red are the computed total energies, while those in black are the computed Gibbs free energies.