Figure 1.
Myonuclei from sedentary mice demonstrate sub-specialized expression of genes key to metabolic regulation
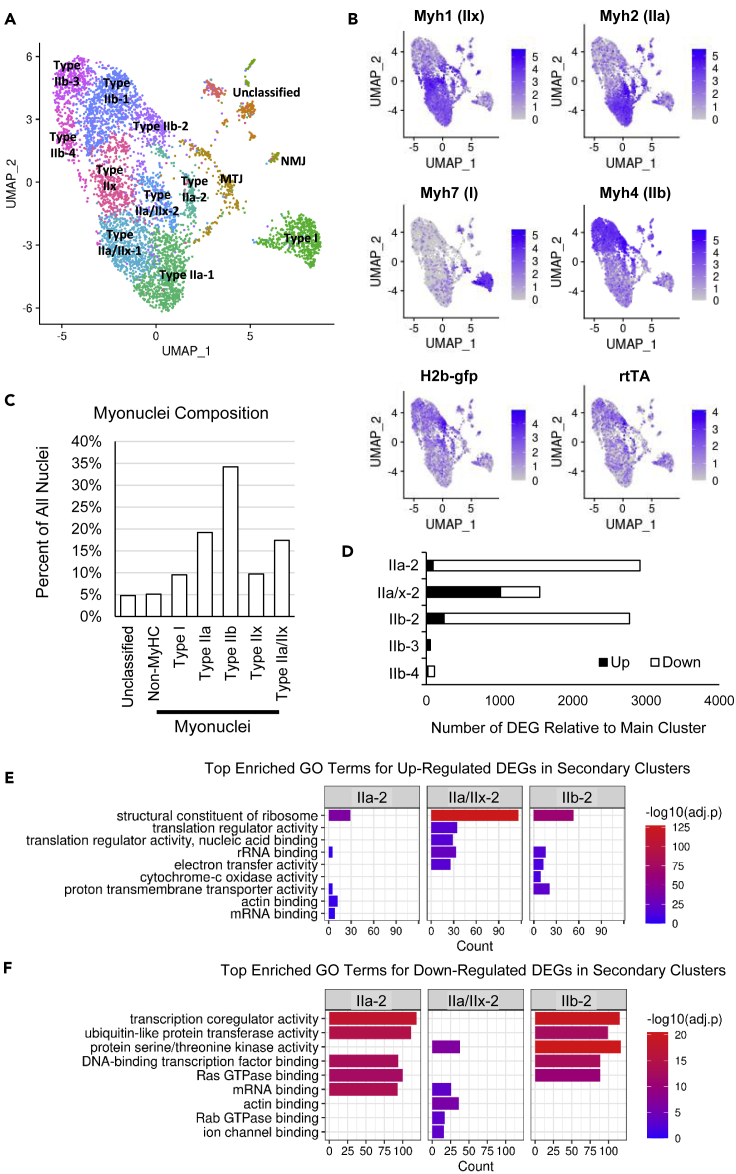
(A) Unbiased clustering and 2-dimensional uniform manifold approximation and projection (UMAP) representation of myonuclei labeled for myosin heavy chain (MyHC) gene expression in soleus and plantaris muscles. MTJ and NMJ represent myo-tendinous junction and neuromuscular junction myonuclei, respectively.
(B) Expression of key genes in post-mitotic myonuclei and precursor cell nuclei including the transgenes for myonuclear labeling. Parenthesis represent the MyHC isoform encoded by the Myh gene.
(C) Nuclei with non-muscle expression profiles compose less than 5% of all nuclei and myonuclei, whereas MyHC as significant gene markers represent around 90% of all nuclei in the sample.
(D) Number of differentially expressed genes (DEGs) when comparing sub-clusters of each type II MyHC-expressing myonuclei with the main cluster (cluster - 1). Although there are four clusters of IIb+ myonuclei, pairwise comparisons of the IIb+ subpopulations (IIb-2, IIb-3, and IIb-4) against the main population (cluster IIb-1) reveals that the IIb-3 and IIb-4 clusters may be artifacts due to few DEGs with no gene ontology (GO) enrichment.
(E) Top enriched GO terms for up-regulated DEGs (black bars in D) shows commonality among the secondary clusters to specialize for components of the ribosome and mitochondria.
(F) Top enriched GO terms for DEGs that are down-regulated (white bars in D) in the secondary clusters.