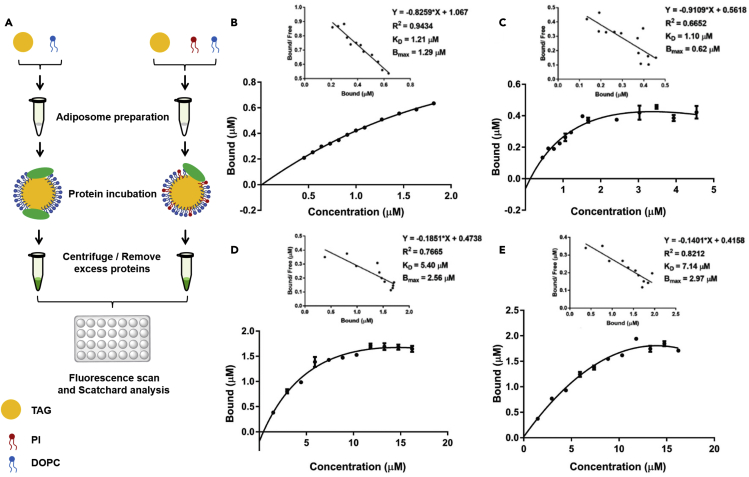
Figure 4.
Reduction of PLIN2 saturated binding on adiposomes by PtdIns
Adiposomes were prepared using a dose of PtdIns and DOPC in a molar ratio. Then, adiposome preparation was aliquoted equally to 30 μl each and incubated with increase doses of PLIN2, supplementing corresponded volume of Tris-NaCl buffer (50 mM Tris-HCl, 150 mM NaCl, pH 7.4) to prepare an equal 60 μl specimen. After incubation, the adiposomes were reisolated and washed to remove nonspecific binding proteins. The fluorescence intensity of adiopsome-bound PLIN2 was determined using an EnSpire Multimode Plate Reader (PerkinElmer) with excitation and emission wavelength at 488 nm and 530 nm for GFP tag, respectively; or 550 nm and 580 nm for APPLE tag, respectively.
(A) Diagram of the experimental design. The Scatchard plot was analyzed using GraphPad 7.0. The xaxis represents the concentration of PLIN2 or PLIN3 in both figures. The concentration of purified proteins was determined by BCA Protein Assay Kit and the fluorescence intensity represented their concentrations on adiposomes. A series of doses of SMT3-PLIN2-GFP and PLIN3-APPLE were incubated with adiposomes at 4°C for 12 h and the fluorescence intensity was detected using EnSpire Multimode Plate Reader.
(B) The saturation curves with Scatchard plot of bound SMT3-PLIN2-GFP on adiposomes with DOPC only.
(C) The saturation curves with Scatchard plots of bound SMT3-PLIN2-GFP on adiposomes with 92.4% DOPC and 7.6% PtdIns, molar ratio.
(D) The saturation curves with Scatchard plots of bound PLIN3-APPLE on adiposomes with DOPC only.
(E) The saturation curves with Scatchard plots of bound PLIN3-APPLE on adiposomes with 92.4% DOPC and 7.6% PtdIns, molar ratio. The equilibrium dissociation constant KD and the maximum saturation concentration of the binding sites on the adiposomes, Bmax, were determined using Scatchard plots. Data represent mean ± s.e.m., n = 3.
See also Figures S2 and S3 and Table S1.