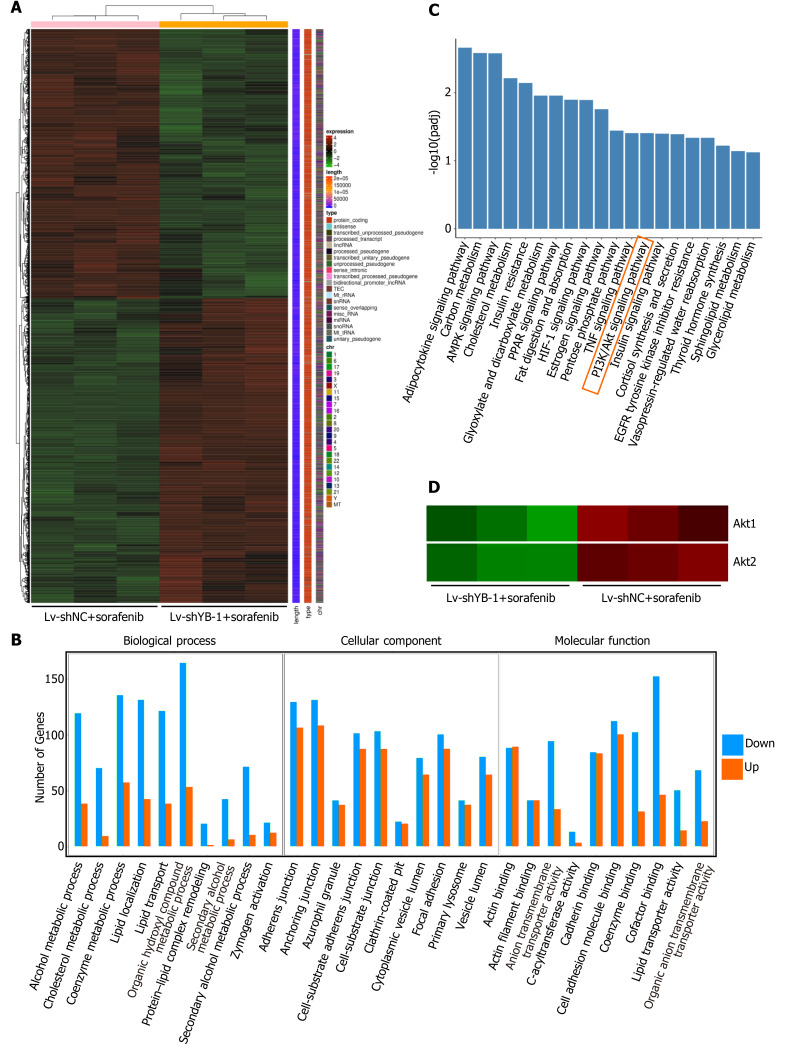
Figure 6.
Digital gene expression profiling-seq analysis in HepG2 cells. A: Heat map of the results from the cluster analysis of the digital gene expression profiling (DGE)-seq data (cells infected with: lentivirus encoding non-specific short hairpin RNA as a negative control (Lv-shNC) + sorafenib vs lentivirus encoding short hairpin RNA against Y box protein 1 (Lv-shYB-1) + sorafenib). Each column denotes the DEGs identified in our study. Each row indicates a sample. For each gene, red represents a high level of expression relative to the mean, while green expresses a low level. The scale bar is the number of standard deviations from the mean; B: Histogram of GO functional analysis for the DEGs obtained from DGE sequencing. GO terms with padj < 0.05 were thought to be notably enriched by DEGs. The y axis represents the number of genes in a GO classification category. Red represents increased expression in the Lv-shYB-1 + sorafenib group, whereas green indicates decreased expression in that group; C: Pathway enrichment analysis showed the top 20 signaling pathways in KEGG (padj < 0.05); D: mRNA expression levels of protein kinase B1 (Akt1) and protein kinase B2 (Akt2) from the DGE-seq data (Lv-shNC + sorafenib, Lv-shYB-1 + sorafenib).