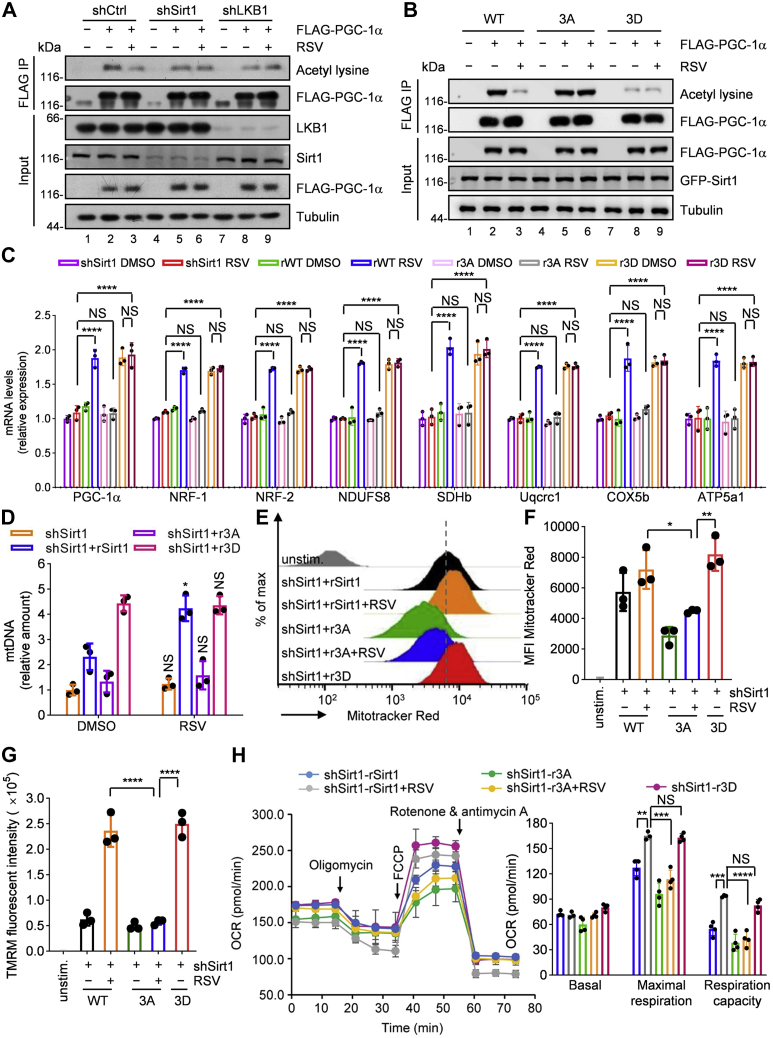
Figure 5.
LKB1-mediated Sirt1 activation promotes mitochondrial function.A, deacetylation of PGC-1α by Sirt1 in resveratrol-treated LKB1-depleted HEK293T cells. HEK293T cells were infected with lentivirus-based particles expressing shRNA control, Sirt1 shRNA, or LKB1 shRNA for 12 h, and 36 h later cells were infected with virus particles expressing FLAG-PGC-1α for 12 h. After 36 h, cells were treated with 25 μM resveratrol for 6 h. FLAG-PGC-1α proteins were immunoprecipitated by anti-FLAG and immunoblotted with anti-acetylated-lysine antibody. Representative of three independent experiments. B, deacetylation of PGC-1α by Sirt1 in resveratrol-treated Sirt1-depleted HEK293T cells expressing WT Sirt1, 3A mutant, or 3D mutant. HEK293T cells stably expressing Sirt1 shRNA were coinfected with lentivirus-based particles expressing FLAG-PGC-1α and lentivirus-based particles expressing GFP-Sirt1 WT, the 3A mutant, or the 3D mutant for 12 h, and 48 h later cells were treated with 25 μM resveratrol for 6 h. FLAG-PGC-1α proteins were immunoprecipitated by anti-FLAG and immunoblotted with anti-acetylated-lysine antibody. C, PGC-1α, NRF-1, NRF-2, NDUFS8, SDHb, Uqcrc1, Cox5b, ATP5a1 mRNA were analyzed by means of quantitative PCR in resveratrol-treated Sirt1-depelted C2C12 cells infected with lentivirus-based particles expressing WT, 3A mutant, or 3D mutant of Sirt1. Relative expression values were normalized to untreated cells. Data represent mean ± SD. The experiment was repeated three times independently. Statistical significance was determined by Tukey's multiple comparisons test. NS (not significant) indicates p > 0.05. ∗∗∗∗p < 0.0001. WT group indicates WT Sirt1 rescue in Sirt1-depleted cells. The 3A group indicates Sirt1 3A mutant (Ser615, Ser669, and Ser732 were replaced by Ala) rescue in Sirt1-depleted cells. The 3D group indicates Sirt1 3D mutant (Ser615, Ser669, and Ser732 were replaced by Asp) rescue in Sirt1-depleted cells. D, mitochondrial content analyzed by means of quantitative PCR in C2C12 cells treated with 25 μM resveratrol. Relative expression values were normalized to untreated cells. Data represent mean ± SD. The experiment was repeated three times independently. Statistical significance was determined by unpaired two-tailed t test. NS (not significant) indicates p > 0.05. The p (shSirt1, shSirt1 RSV) = 0.2497, ∗p (shSirt1+rSirt1, shSirt1+rSirt1 RSV) = 0.0103, p (shSirt1+r3A, shSirt1+r3A RSV) = 0.5695, p (shSirt1+r3D, shSirt1+r3D RSV) = 0.8046. The shSirt1+rSirt1 group indicates WT Sirt1 rescue in Sirt1-depleted cells. The shSirt1+r3A group indicates Sirt1 3A mutant rescue in Sirt1-depleted cells. The shSirt1+r3D group indicates Sirt1 3D mutant rescue in Sirt1-depleted cells. E, mitochondrial density in C2C12 cells was measured by flow cytometry using MitoTracker Red CMXRos. F, the relative change in the mean fluorescence intensity of MitoTracker in panel E. Data represent mean ± SD. The experiment was repeated three times independently. Statistical significance was determined by Tukey's multiple comparisons test. ∗p = 0.0252, ∗∗p = 0.0026. G, in C2C12 cells used in panel E, mitochondrial membrane potential was measured by ImageStream mark ii flow cytometer using tetramethyl rhodamine methyl ester (TMRM). Data represent mean ± SD. The experiment was repeated three times independently. Statistical significance was determined by Tukey's multiple comparisons test. ∗∗∗∗p < 0.0001. H, in C2C12 cells used in panel E, seahorse assays were conducted. Oxygen consumption rate (OCR) over time (left panel) and OCR in different stages of the measurement (right panel) are shown. Data represent mean ± SD. Statistical significance was determined by Tukey's multiple comparisons test. NS (not significant) indicates p > 0.05. In maximal respiration measurement, ∗∗p = 0.0026, ∗∗∗p = 0.0001. In respiration capacity measurement, ∗∗∗p = 0.0001, ∗∗∗∗p < 0.0001. RSV, resveratrol.