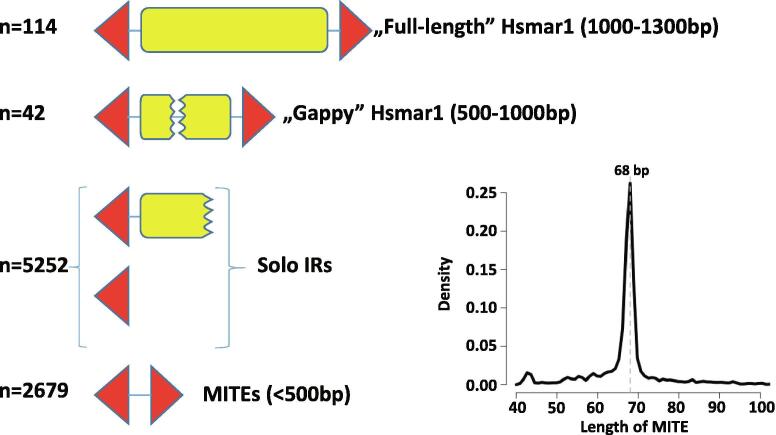
Fig. 1.
Characteristics of Hsmar1 mariner transposons. “Full-length” Hsmar1, “gappy” Hsmar1, and MITEs (Miniature Inverted Repeat Transposable Elements) contain two inverted terminal repeat (ITR) sequences (red triangles). “Solo” inverted repeats contain one ITR linked to a truncated Hsmar1 sequence or not. In each category, up to three mismatches were allowed in the flanking ITR sequences (“core” motif: 5’-GGTGCAAAAGTAATTGCGG-3’). Histogram shows the length distribution of MITE sequences with a median size of 68 bp. Only ITRs within 500 bp were considered to identify MITEs. The number of cases (n) is shown on the left. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)